Key Points
-
General introduction to bacterial membrane protein insertion and the Sec translocases.
-
Membrane protein topology and targeting to the membrane.
-
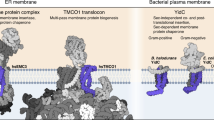
'YidC-only' pathway for bacterial membrane protein insertion.
-
YidC family members and their structure function.
-
Bacterial Sec pathway for membrane protein insertion and comparison with the eukaryotic Sec system in the endoplasmic reticulum.
-
SecYEG structure.
-
Folding, assembly and quality control of bacterial membrane proteins.
-
Conclusion: resolved problems and open questions.
Abstract
This Review describes the pathways that are used to insert newly synthesized proteins into the cytoplasmic membranes of bacteria, and provides insight into the function of two of the evolutionarily conserved translocases that catalyse this process. These highly sophisticated translocases are responsible for decoding the topogenic sequences within membrane proteins that direct membrane protein insertion and orientation. The role of the Sec and YidC translocases in the folding of bacterial membrane proteins is also highlighted.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
11 December 2017
This article was published with an incorrect DOI. The correct DOI is 10.1038/nrmicro3595. This has now been corrected in the online version. We apologize to the authors and to readers for any confusion caused.
References
Daley, D. O. et al. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308, 1321–1323 (2005).
Casadio, R., Fariselli, P., Finocchiaro, G. & Martelli, P. L. Fishing new proteins in the twilight zone of genomes: the test case of outer membrane proteins in Escherichia coli K12, Escherichia coli O157:H7, and other Gram-negative bacteria. Protein Sci. 12, 1158–1168 (2003).
Ito, K. & Akiyama, Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59, 211–231 (2005).
Shimohata, N., Nagamori, S., Akiyama, Y., Kaback, H. R. & Ito, K. SecY alterations that impair membrane protein folding and generate a membrane stress. J. Cell Biol. 176, 307–317 (2007).
Tokuda, H. & Matsuyama, S. Sorting of lipoproteins to the outer membrane in E. coli . Biochim. Biophys. Acta 1693, 5–13 (2004).
Dalbey, R. E. & Chen, M. Sec-translocase mediated membrane protein biogenesis. Biochim. Biophys. Acta 1694, 37–53 (2004).
Luirink, J., von Heijne, G., Houben, E. & de Gier, J. W. Biogenesis of inner membrane proteins in Escherichia coli . Annu. Rev. Microbiol. 59, 329–355 (2005).
Muller, M. & Klosgen, R. B. The Tat pathway in bacteria and chloroplasts. Mol. Membr. Biol. 22, 113–121 (2005).
Lee, P. A., Tullman-Ercek, D. & Georgiou, G. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60, 373–395 (2006).
Hatzixanthis, K., Palmer, T. & Sargent, F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 49, 1377–1390 (2003).
Yi, L. & Dalbey, R. E. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria. Mol. Membr. Biol. 22, 101–111 (2005).
Kiefer, D. & Kuhn, A. YidC as an essential and multifunctional component in membrane protein assembly. Int. Rev. Cytol. 259, 113–138 (2007).
Hardy, S. J. & Randall, L. L. Recognition of ligands by SecB, a molecular chaperone involved in bacterial protein export. Philos. Trans. R. Soc. Lond. B 339, 343–352; discussion 352–354 (1993).
Ulbrandt, N. D., Newitt, J. A. & Bernstein, H. D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196 (1997).
Macfarlane, J. & Muller, M. The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem. 233, 766–771. (1995).
Schierle, C. F., Berkmen, M., Huber, D., Kumamoto, C., Boyd, D. & Beckwith, J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185, 5706–5713 (2003).
Bowers, C. W., Lau, F. & Silhavy, T. J. Secretion of LamB–LacZ by the signal recognition particle pathway of Escherichia coli . J. Bacteriol. 185, 5697–5705 (2003).
Raine, A. et al. Targeting and insertion of heterologous membrane proteins in E. coli . Biochimie 85, 659–668 (2003).
Luirink, J. & Sinning, I. SRP-mediated protein targeting: structure and function revisited. Biochim. Biophys. Acta 1694, 17–35 (2004). A useful report on how proteins are targeted to the membrane by the SRP pathway in the three domains of life.
Valent, Q. A. et al. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol. Microbiol. 25, 53–64 (1997).
Herskovits, A. A. et al. Evidence for coupling of membrane targeting and function of the signal recognition particle (SRP) receptor FtsY. EMBO Rep. 2, 1040–1046 (2001).
Facey, S. J., Neugebauer, S. A., Krauss, S. & Kuhn, A. The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli . J. Mol. Biol. 365, 995–1004 (2007).
Gallusser, A. & Kuhn, A. Initial steps in protein membrane insertion. Bacteriophage M13 procoat protein binds to the membrane surface by electrostatic interaction. EMBO J. 9, 2723–2729 (1990).
de Gier, J. W. et al. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli . Proc. Natl Acad. Sci. USA 95, 14646–14651 (1998).
de Gier, J. W. et al. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399, 307–309 (1996).
Scotti, P. A. et al. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 (2000). Showed that a portion of YidC co-purifies with the Sec translocase and that YidC makes contact with the TM region of a membrane protein during insertion.
Stuart, R. A. & Neupert, W. Making membranes in bacteria. Nature 406, 575–577 (2000).
Samuelson, J. C. et al. YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 (2000).
Samuelson, J. C. et al. Function of YidC for the insertion of M13 procoat protein in E. coli: translocation of mutants that show differences in their membrane potential dependence and Sec-requirement. J. Biol. Chem. 276, 34847–34852 (2001).
Chen, M. et al. Direct interaction of YidC with the Sec-independent Pf3 coat protein during its membrane protein insertion. J. Biol. Chem. 277, 7670–7675 (2002).
Geller, B. L. & Wickner, W. M13 procoat inserts into liposomes in the absence of other membrane proteins. J. Biol. Chem. 260, 13281–13285 (1985).
Serek, J. et al. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23, 294–301 (2004).
Van Der Laan, M. et al. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl Acad. Sci. USA 100, 5801–5806 (2003).
Yi, L., Celebi, N., Chen, M. & Dalbey, R. E. Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279, 39260–39267 (2004).
van Bloois, E., Jan Haan, G., de Gier, J. W., Oudega, B. & Luirink, J. F1F0 ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 576, 97–100 (2004).
Celebi, N., Yi, L., Facey, S. J., Kuhn, A. & Dalbey, R. E. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357, 1428–1436 (2006).
van Bloois, E., Haan, G. J., de Gier, J. W., Oudega, B. & Luirink, J. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281, 10002–10009 (2006).
du Plessis, D. J., Nouwen, N. & Driessen, A. J. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281, 12248–12252 (2006).
van Der Laan, M., Bechtluft, P., Kol, S., Nouwen, N. & Driessen, A. J. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222 (2004). References 32 and 39 show that YidC is sufficient to promote the membrane insertion of Sec-independent proteins.
Yen, M. R., Harley, K. T., Tseng, Y. H. & Saier, M. H. Jr. Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol. Lett. 204, 223–231 (2001).
Luirink, J., Samuelsson, T. & de Gier, J. W. YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501, 1–5 (2001).
Jiang, F. et al. Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J. Biol. Chem. 277, 19281–19288 (2002).
van Bloois, E. et al. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J. Biol. Chem. 280, 12996–13003 (2005).
Preuss, M., Ott, M., Funes, S., Luirink, J. & Herrmann, J. M. Evolution of mitochondrial Oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J. Biol. Chem. 280, 13004–13011 (2005).
Jia, L. et al. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22, 6438–6447 (2003).
Szyrach, G., Ott, M., Bonnefoy, N., Neupert, W. & Herrmann, J. M. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22, 6448–6457 (2003).
Tjalsma, H., Bron, S. & van Dijl, J. M. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278, 15622–15632 (2003).
Hasona, A. et al. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl Acad. Sci. USA 102, 17466–17471 (2005).
Bender, G. R., Sutton, S. V. & Marquis, R. E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 53, 331–338 (1986).
Nouwen, N. & Driessen, A. J. SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44, 1397–1405 (2002).
Xie, K., Kiefer, D., Nagler, G., Dalbey, R. E. & Kuhn, A. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45, 13401–13408 (2006).
Oliver, D. C. & Paetzel, M. Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J. Biol. Chem. 19 Dec 2007 (doi:10.1074/jbc.M708936200).
Jiang, F. et al. Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 278, 48965–48972 (2003).
Nargang, F. E., Preuss, M., Neupert, W. & Herrmann, J. M. The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa . J. Biol. Chem. 277, 12846–12853 (2002).
Veenendaal, A. K., van der Does, C. & Driessen, A. J. The protein-conducting channel SecYEG. Biochim. Biophys. Acta 1694, 81–95 (2004).
Papanikou, E., Karamanou, S. & Economou, A. Bacterial protein secretion through the translocase nanomachine. Nature Rev. Microbiol. 5, 839–851 (2007).
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004). A landmark paper on the structure of the Sec translocation channel in archaea.
Pogliano, J. A. & Beckwith, J. SecD and SecF facilitate protein export in Escherichia coli . EMBO J. 13, 554–561 (1994).
Chen, M. et al. Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 44, 10741–10749 (2005).
Vrontou, E. & Economou, A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim. Biophys. Acta 1694, 67–80 (2004).
Gelis, I. et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131, 756–769 (2007).
Kuhn, A. Alterations in the extracellular domain of M13 procoat protein make its membrane insertion dependent on secA and secY . Eur. J. Biochem. 177, 267–271 (1988).
Andersson, H. & von Heijne, G. Sec dependent and Sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J. 12, 683–691 (1993).
Cao, G., Kuhn, A. & Dalbey, R. E. The translocation of negatively charged residues across the membrane is driven by the electrochemical potential: evidence for an electrophoresis-like membrane transfer mechanism. EMBO J. 14, 866–875 (1995).
Deitermann, S., Sprie, G. S. & Koch, H. G. A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli . J. Biol. Chem. 280, 39077–39085 (2005).
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005). This paper reports the features of a polypeptide segment that promotes lateral release from the Sec translocon into the ER membrane.
Kuhn, A., Stuart, R., Henry, R. & Dalbey, R. E. The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13, 510–516 (2003).
Houben, E. N., ten Hagen-Jongman, C. M., Brunner, J., Oudega, B. & Luirink, J. The two membrane segments of leader peptidase partition one by one into the lipid bilayer via a Sec/YidC interface. EMBO Rep. 5, 970–975 (2004).
Beck, K. et al. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714 (2001).
Cannon, K. S., Or, E., Clemons, W. M. Jr, Shibata, Y. & Rapoport, T. A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 169, 219–225 (2005).
Maillard, A. P., Lalani, S., Silva, F., Belin, D. & Duong, F. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J. Biol. Chem. 282, 1281–1287 (2007).
Tam, P. C., Maillard, A. P., Chan, K. K. & Duong, F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 24, 3380–3388 (2005).
Harris, C. R. & Silhavy, T. J. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 181, 3438–3444 (1999).
Plath, K., Mothes, W., Wilkinson, B. M., Stirling, C. J. & Rapoport, T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807 (1998).
Hanein, D. et al. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell 87, 721–732 (1996).
Osborne, A. R. & Rapoport, T. A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 129, 97–110 (2007).
Mitra, K. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005).
Beckmann, R. et al. Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science 278, 2123–2126 (1997).
Crowley, K. S., Liao, S., Worrell, V. E., Reinhart, G. D. & Johnson, A. E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461–471 (1994).
von Heijne, G. Recent advances in the understanding of membrane protein assembly and structure. Q. Rev. Biophys. 32, 285–307 (1999).
Popot, J. L. & Engelman, D. M. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69, 881–922 (2000).
Nagamori, S., Smirnova, I. N. & Kaback, H. R. Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165, 53–62 (2004).
Stenberg, F., von Heijne, G. & Daley, D. O. Assembly of the cytochrome bo 3 complex. J. Mol. Biol. 371, 765–773 (2007).
Kulajta, C., Thumfart, J. O., Haid, S., Daldal, F. & Koch, H. G. Multi-step assembly pathway of the cbb 3 -type cytochrome c oxidase complex. J. Mol. Biol. 355, 989–1004 (2006).
Hiser, L. & Hosler, J. P. Heme A is not essential for assembly of the subunits of cytochrome c oxidase of Rhodobacter sphaeroides . J. Biol. Chem. 276, 45403–45407 (2001).
Nijtmans, L. G., Taanman, J. W., Muijsers, A. O., Speijer, D. & Van den Bogert, C. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 254, 389–394 (1998).
Wielburski, A. & Nelson, B. D. Heme a induces assembly of rat liver cytochrome c oxidase subunits I–III in isolated mitochondria. FEBS Lett. 177, 291–294 (1984).
Kihara, A., Akiyama, Y. & Ito, K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl Acad. Sci. USA 92, 4532–4536 (1995).
Akiyama, Y., Kihara, A. & Ito, K. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli . FEBS Lett. 399, 26–28 (1996).
Kihara, A., Akiyama, Y. & Ito, K. Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18, 2970–2981 (1999).
Akiyama, Y., Kanehara, K. & Ito, K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 23, 4434–4442 (2004).
Sakoh, M., Ito, K. & Akiyama, Y. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli . J. Biol. Chem. 280, 33305–33310 (2005).
Maegawa, S., Ito, K. & Akiyama, Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry 44, 13543–13552 (2005).
Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 (2003).
Paschen, S. A. et al. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 (2003).
Gentle, I., Gabriel, K., Beech, P., Waller, R. & Lithgow, T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 (2004).
Kozjak, V. et al. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278, 48520–48523 (2003).
Bolter, B., Soll, J., Schulz, A., Hinnah, S. & Wagner, R. Origin of a chloroplast protein importer. Proc. Natl Acad. Sci. USA 95, 15831–15836 (1998).
Ruiz, N., Falcone, B., Kahne, D. & Silhavy, T. J. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317 (2005).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli . Cell 121, 235–245 (2005).
Sklar, J. G. et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli . Proc. Natl Acad. Sci. USA 104, 6400–6405 (2007).
Kim, S. et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007).
Clantin, B. et al. Structure of the membrane protein FhaC: a member of the Omp85–TpsB transporter superfamily. Science 317, 957–961 (2007).
Robert, V. et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4, e377 (2006).
Habib, S. J. et al. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 176, 77–88 (2007).
Rapoport, T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 (2007).
Yuan, J., Henry, R., McCaffery, M. & Cline, K. SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science 266, 796–798 (1994).
Laidler, V., Chaddock, A. M., Knott, T. G., Walker, D. & Robinson, C. A SecY homolog in Arabidopsis thaliana. Sequence of a full-length cDNA clone and import of the precursor protein into chloroplasts. J. Biol. Chem. 270, 17664–17667 (1995).
Schuenemann, D., Amin, P., Hartmann, E. & Hoffman, N. E. Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa but not the 23-kDa subunit of the oxygen-evolving complex. J. Biol. Chem. 274, 12177–12182 (1999).
Hartmann, E. et al. Evolutionary conservation of components of the protein translocation complex. Nature 367, 654–657 (1994).
Rapoport, T. A., Jungnickel, B. & Kutay, U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65, 271–303 (1996).
Ring, G. & Eichler, J. Extreme secretion: protein translocation across the archael plasma membrane. J. Bioenerg. Biomembr. 36, 35–45 (2004).
Pohlschroder, M., Hartmann, E., Hand, N. J., Dilks, K. & Haddad, A. Diversity and evolution of protein translocation. Annu. Rev. Microbiol. 59, 91–111 (2005).
Goder, V. & Spiess, M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 22, 3645–3653 (2003).
Liao, S., Lin, J., Do, H. & Johnson, A. E. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell 90, 31–41 (1997).
Woolhead, C. A., McCormick, P. J. & Johnson, A. E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 (2004).
Lu, J. & Deutsch, C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry 44, 8230–8243 (2005).
Lu, J. & Deutsch, C. Folding zones inside the ribosomal exit tunnel. Nature Struct. Mol. Biol. 12, 1123–1129 (2005).
Do, H., Falcone, D., Lin, J., Andrews, D. W. & Johnson, A. E. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell 85, 369–378 (1996).
Sadlish, H., Pitonzo, D., Johnson, A. E. & Skach, W. R. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nature Struct. Mol. Biol. 12, 870–878 (2005).
Acknowledgements
Work in the laboratory of R.E.D. was supported by National Institutes of Health Grant GM63862-05. The authors thank A. Kuhn for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Entrez Genome Project
Entrez Protein
FURTHER INFORMATION
Glossary
- Type I membrane protein
-
A protein which contains a single membrane-spanning domain that has its carboxyl terminus orientated towards the cytoplasm and its amino terminus orientated towards the lumen of membrane compartments or in an extracellular direction.
- Type II membrane protein
-
A single-spanning membrane protein that has the opposite topology to a type I membrane protein.
- Signal-recognition particle
-
A complex that is responsible for targeting nascent polypeptides to the cell membranes, and identifies an amino-terminal signal sequence that is carried by proteins that are destined for secretion or membrane localization.
- Two-partner secretion system
-
A secretion system that is composed of two distinct proteins; one is secreted and the other is its transporter.
- Signal anchor
-
A topogenic sequence that signals the initiation of translocation of the carboxy-terminal region of a membrane protein, and remains as a membrane anchor with an NinCout orientation. Also known as a type II signal anchor.
- Reverse signal anchor
-
A topogenic sequence that signals the initiation of translocation of the amino-terminal region of a membrane protein, and remains as a membrane-spanning region that has an NoutCin orientation. Also known as a type I signal anchor.
- Signal peptidase II
-
A signal peptidase that proteolytically removes lipoprotein signal sequences.
Rights and permissions
About this article
Cite this article
Xie, K., Dalbey, R. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol 6, 234–244 (2008). https://doi.org/10.1038/nrmicro3595
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3595
This article is cited by
-
Erratum: Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases
Nature Reviews Microbiology (2018)
-
MifM-instructed translation arrest involves nascent chain interactions with the exterior as well as the interior of the ribosome
Scientific Reports (2018)
-
Protein export through the bacterial Sec pathway
Nature Reviews Microbiology (2017)
-
In vitro membrane protein synthesis inside Sec translocon-reconstituted cell-sized liposomes
Scientific Reports (2016)
-
Identifying co-targets to fight drug resistance based on a random walk model
BMC Systems Biology (2012)