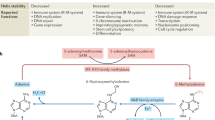
Key Points
-
N6-methyl-adenine is a common DNA modification in bacterial genomes, which is catalysed by two classes of DNA adenine methyltransferases: those associated with restriction–modification (R–M) systems and 'solitary' methyltransferases that do not have a restriction-enzyme companion.
-
R–M systems protect bacteria from the invasion of foreign DNA (for example, phages). Each R–M system is made up of a restriction enzyme and a modification enzyme, which both recognize the same DNA target. Some modification enzymes are DNA adenine methyltransferases, whereas others are DNA cytosine methyltransferases.
-
In γ-proteobacteria, methylation of the adenine moiety at GATC sites by the Dam methylase provides signals for chromosome replication, nucleoid organization and segregation, mismatch repair, transposition of insertion elements, phase variation, bacterial conjugation and packaging of phage DNA. Furthermore, Dam methylation is required for virulence in Salmonella, Haemophilus, Yersinia and Vibrio species.
-
In uropathogenic Escherichia coli, phase variation in Pap fimbriae is regulated at the transcriptional level by Dam methylation and the leucine-responsive regulatory protein (Lrp). Synthesis of Prf, S, Afa, K88 and CS31a fimbriae is also regulated by Dam methylation and Lrp. In turn, phase variation in the E. coli Agn43 antigen is regulated by Dam and OxyR.
-
In Salmonella, Dam methylation regulates the expression of genes involved in invasion of epithelial cells (SPI-1 genes), synthesis of fimbrial adhesins (Pef and Std), envelope proteins (Braun lipoprotein), flagella and chemotaxis. Also, Dam methylation is required for bile resistance.
-
Dam methylation regulates conjugal transfer of the Salmonella virulence plasmid and other F-like plasmids.
-
In Caulobacter and other α-proteobacteria, methylation of the adenine moiety at GANTC sites by the CcrM methylase regulates the cell cycle and is essential for viability. Lack of CcrM methylation attenuates virulence in Brucella.
-
DNA adenine methylation is found in the genomes of certain protists and fungi, and might exist in other eukaryotes.
Abstract
N6-methyl-adenine is found in the genomes of bacteria, archaea, protists and fungi. Most bacterial DNA adenine methyltransferases are part of restriction–modification systems. Certain groups of Proteobacteria also harbour solitary DNA adenine methyltransferases that provide signals for DNA–protein interactions. In γ-proteobacteria, Dam methylation regulates chromosome replication, nucleoid segregation, DNA repair, transposition of insertion elements and transcription of specific genes. In Salmonella, Haemophilus, Yersinia and Vibrio species and in pathogenic Escherichia coli, Dam methylation is required for virulence. In α-proteobacteria, CcrM methylation regulates the cell cycle in Caulobacter, Rhizobium and Agrobacterium, and has a role in Brucella abortus infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cheng, X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 24, 293–318 (1995).
Barbeyron, T., Kean, K. & Forterre, P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J. Bacteriol. 160, 586–590 (1984).
Bickle, T. A. & Kruger, D. H. Biology of DNA restriction. Microbiol. Rev. 57, 434–450 (1993).
Marinus, M. G. Methylation of DNA. In Escherichia coli and Salmonella: Cellular and Molecular Biology (eds Neidhardt, F. C. et al.) 782–791 (ASM Press, Washington DC, 1996). A comprehensive summary of the first two decades of research on Dam methylation.
Reisenauer, A., Kahng, L. S., McCollum, S. & Shapiro, L. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181, 5135–5139 (1999).
Low, D. A., Weyand, N. J. & Mahan, M. J. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69, 7197–7204 (2001). The only published survey of the roles of Dam methylation in bacterial virulence.
Løbner-Olesen, A., Skovgaard, O. & Marinus, M. Dam methylation: coordinating cellular processes. Curr. Op. Microbiol. 8, 154–160 (2005). An update on the roles of Dam methylation in bacteria, including evolutionary aspects.
Esteller, M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 45, 629–656 (2005).
Engel, J. D. & von Hippel, P. H. Effects of methylation on the stability of nucleic acid conformation: studies at the polymer level. J. Biol. Chem. 253, 928–934 (1978).
Diekmann, S. DNA methylation can enhance or induce DNA curvature. EMBO J. 6, 4213–4217 (1987).
Polaczek, P., Kwan, K. & Campbell, J. L. GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA–protein recognition. Mol. Gen. Genet. 258, 488–493 (1998).
Messer, W. & Noyer-Weidner, M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell 54, 735–737 (1988).
Luria, S. E. & Human, M. L. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 64, 557–569 (1952).
Bertani, G. & Weigle, J. J. Host controlled variation in bacterial viruses. J. Bacteriol. 65, 113–121 (1953).
Arber, W. & Dussoix, D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage λ. J. Mol. Biol. 5, 18–36 (1962).
Lacks, S. & Greenberg, B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol. 114, 153–168 (1977).
Murray, N. E. Immigration control of DNA in bacteria: self versus non-self. Microbiology 148, 3–20 (2002). A comprehensive review on the biological significance of R–M systems.
McKane, M. & Milkman, R. Transduction, restriction and recombination patterns in Escherichia coli. Genetics 139, 35–43 (1995).
Jeltsch, A. Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/modification systems? Gene 317, 13–16 (2003).
Kobayashi, I. Behavior of restriction–modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29, 3742–3756 (2001).
Urig, S. et al. The Escherichia coli Dam DNA methyltransferase modifies DNA in a highly processive reaction. J. Mol. Biol. 319, 1085–1096 (2002).
Torreblanca, J. & Casadesús, J. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics 144, 15–26 (1996).
Julio, S. M. et al. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia enterocolitica and Vibrio cholerae. Infect. Immun. 69, 7610–7615 (2001).
Egan. E. S. & Waldor M. K. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114, 521–530.
Boye, E., Løbner-Olesen, A. & Skarstad, K. Limiting DNA replication to once and only once. EMBO Rep. 1, 479–483 (2000).
Campbell, J. L. & Kleckner, N. The E. coli oriC and the dnaA gene promoter are sequestered from Dam methyltransferase following the passage of the chromosomal replication fork. Cell 62, 967–979 (1990).
Lu, M., Campbell, J. L., Boye, E. & Kleckner, N. SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426 (1994).
von Friesleben, U., Rasmussen, K. V. & Schaechter, M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14, 763–772 (1994).
Boye, E., Stokke, T., Kleckner, N. & Skarstad, K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA 93, 12206–12211 (1996).
Braun, R. W. & Wright, A. DNA methylation differentially enhances the expression of one of the two dnaA promoters in vivo and in vitro. Mol. Gen. Genet. 202, 246–250 (1986).
Kücherer, C., Lother, H., Kölling, R., Schauzu, M. A. & Messer, W. Regulation of transcription of the chromosomal dnaA gene of Escherichia coli. Mol. Gen. Genet. 205, 115–121 (1986).
Riber, L. & Løbner-Olesen, A. Coordinated replication and sequestration of oriC and dnaA are required for maintaining once-per-cell-cycle initiation in Escherichia coli. J. Bacteriol. 187, 5605–5613 (2005).
d'Alençon, E et al. Isolation of a new hemimethylated DNA binding protein which regulates dnaA gene expression. J. Bacteriol. 185, 2967–2971 (2003).
Ogden, G. B., Pratt, M. J. & Schaechter, M. The replicative origin of the E. coli chromosome binds to cell membranes only when heminethylated. Cell 54, 121–135 (1988).
Herrick, J. et al. Parental strand recognition of the DNA replication origin by the outer membrane in Escherichia coli. EMBO J. 13, 4695–4703 (1994).
Bach, T., Krekling, M. A. & Skarstad, K. Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J. 22, 315–323 (2003).
Brendler, T., Sawitzke, J., Sergueev, K. & Austin, S. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 19, 6249–6258 (2000).
Løbner-Olesen, A., Marinus, M. G. & Hansen, F. G. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl Acad. Sci. USA 100, 4672–4677 (2003).
Yamazoe, M., Adachi, S., Kanava, S., Obsumi, K. & Hiraga, S. Sequential binding of SeqA protein to nascent DNA segments at replication forks in synchronized cultures of E. coli. Mol. Microbiol. 55, 289–298 (2005).
Hsieh, T. Molecular mechanisms of DNA mismatch repair. Mutat. Res. 486, 71–87 (2001).
Burdett, V., Baitinger, C., Visvanathan, C., Lovett, S. & Modrich, P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA 98, 6765–6770 (2001).
Welsch, K. M., Lu, A. L., Clark, S. & Modrich, P. Isolation and characterization of the E. coli mutH product. J. Biol. Chem. 262, 15624–15629 (1987).
Glickman, B. W. & Radman, M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl Acad. Sci. USA 77, 1063–1067 (1980).
Fram, R. J., Cusick, P. S., Wilson, J. M. & Marinus, M. G. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol. Pharmacol. 28, 51–55 (1985).
Karran, P. & Marinus, M. G. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature 296, 868–869 (1982).
Prieto, A. I., Ramos-Morales, F. & Casadesús, J. Bile-induced DNA damage in Salmonella enterica. Genetics 168, 1787–1794 (2004).
Rasmussen, L. J., Løbner-Olesen, A. & Marinus, M. G. Growth-rate-dependent transcription initiation from the dam P2 promoter. Gene 157, 213–215 (1995).
Roberts, D., Hoopes, B. C., McClure, W. R. & Kleckner, N. IS 10 transposition is regulated by DNA adenine methylation. Cell 43, 117–130 (1985). A classic, elegant study of DNA–protein interactions regulated by m6A.
Yin, J. C., Krebs, M. P. & Reznikoff, W. S. Effect of Dam methylation on Tn5 transposition. J. Mol. Biol. 199, 35–45 (1988).
Dodson, K. W. & Berg, D. E. Factors affecting transposition activity of IS50 and Tn5 ends. Gene 76, 207–213 (1989).
van der Woude, M., Braaten, B. & Low, D. A. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4, 5–9 (1996).
Hernday, A., Krabbe, M., Braaten, B. & Low, D. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl Acad. Sci. USA 99, 16570–16476 (2002). A detailed description of the molecular mechanisms that control the pap operon, a paradigm among bacterial epigenetic switches.
Hernday, A. D., Braaten B. A. & Low, D. A. The mechanism by which DNA adenine methylase and PapI activate the Pap epigenetic switch. Mol. Cell 12, 947–957 (2003).
White-Ziegler, C. A, Black, A. M., Eliades, S. H., Young, S. & Porter, K. The N-acetyltransferase RimJ responds to environmental stimuli to repress pap fimbrial transcription in Escherichia coli. J. Bacteriol. 184, 4334–4342 (2002).
Hernday, A. D., Braaten, B. A., Broitman-Maduro, G., Engelberts, P. & Low, D. A. Regulation of the Pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16, 537–547 (2004).
Nicholson, B. & Low, D. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35, 728–742 (2000).
Waldron, D. E., Owen, P. & Dorman, C. J. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44, 509–520 (2002).
Haagmans, W. & van der Woude, M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35, 877–887 (2000).
Henderson, L. R. & Owen, P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181, 2132–2141 (1999).
Correnti, J., Munster, M., Chan, T. & van der Woude, M. Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol. Microbiol. 44, 521–532 (2002).
Wallecha, A., Munster, V., Correnti, J., Chan, T. & van der Woude, M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184, 3338–3347 (2002).
Hale, W. B., van der Woude, M. W. & Low, D. A. Analysis of non-methylated GATC sites in the Escherichia coli chromosome and identification of sites that are differentially methylated in response to environmental stimuli. J. Bacteriol. 176, 3438–3441 (1994).
van der Woude, M., Hale, W. B. & Low, D. A. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J. Bacteriol. 180, 5913–5920 (1998).
Mashhoon, N. et al. Functional characterization of Escherichia coli DNA adenine methyltransferase, a novel target for antibiotics. J. Biol. Chem. 279, 52075–52081 (2004).
Torreblanca, J., Marqués, S. & Casadesús, J. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152, 31–45 (1999).
Camacho, E. M. & Casadesús, J. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44, 1589–1598 (2002).
Camacho, E. M. et al. Regulation of finP transcription by DNA adenine methylation in the virulence plasmid of Salmonella enterica. J. Bacteriol. 187, 5691–5699 (2005).
Camacho, E. M. & Casadesús, J. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 57, 1700–1718 (2005).
Heithoff, D. M., Sinsheimer, R. L., Low, D. A. & Mahan, M. J. An essential role for DNA adenine methylation in bacterial virulence. Science 284, 967–970 (1999).
García-del Portillo, F., Pucciarelli, M. G. & Casadesús, J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl Acad. Sci. USA 96, 11578–11583 (1999).
Giacomodonato, M. N., Sarnacki, M. H., Caccuri, R. L., Sordelli, D. O. & Cerquetti, M. C. Host response to a dam mutant of Salmonella enterica serovar Enteritidis with a temperature sensitive phenotype. Infect. Immun. 72, 5498–5501 (2004).
Pucciarelli, M. G., Prieto, A. I., Casadesús, J. & García-del Portillo, F. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148, 1171–1182 (2002).
Heithoff, D. M. et al. Salmonella DNA adenine methylase mutants confer cross-protective immunity. Infect. Immun. 69, 6725–6730 (2001).
Honma, Y., Fernández, R. E. & Maurelli, A. T. A DNA adenine methylase mutant of Shigella flexneri shows no significant attenuation of virulence. Microbiology 150, 1073–1078 (2004).
Dueger, E. L., House, J. K., Heithoff, D. M. & Mahan, M. J. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine 21, 3249–3258 (2003).
Watson, M. E. Jr ., Jarisch, J. & Smith, A. L. Inactivation of deoxyadenosine methyltransferase (dam) attenuates Haemophilus influenzae virulence. Mol. Microbiol. 53, 651–654 (2004).
Taylor, V. L., Titball, R. W. & Oyston, P. C. F. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects again plague. Microbiology 151, 1919–1926 (2005).
Julio, S. M., Heithoff, D. M., Sinsheimer, R. L., Low, D. A. & Mahan, M. J. DNA adenine methylase overproduction in Yersinia pseudotuberculosis alters YopE expression and secretion and host immune responses to infection. Infect. Immun. 70, 1006–1009 (2002).
Fälker, S., Schmidt, M. A. & Heusipp, G. DNA methylation in Yersinia enterocolitica: role of the DNA adenine methyltransferase in mismatch repair and regulation of virulence factors. Microbiology 151, 2291–2299 (2005).
Chen, L. et al. Alteration of DNA adenine methylase (Dam) activity in Pasteurella multocida causes increased spontaneous mutation frequency and attenuation in mice. Microbiology 149, 2283–2290 (2003).
Oshima, T. et al. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45, 673–695 (2002).
McClelland, M. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J. Mol. Evol. 21, 317–322 (1985).
Blaisdell, B. E., Campbell, A. M. & Karlin, S. Similarities and dissimilarities of phage genomes. Proc. Natl Acad. Sci. USA 93, 5854–5859 (1996).
Sternberg, N., Sauer, B., Hoess, R. & Abremski, K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by Dam methylation. J. Mol. Biol. 187, 197–212 (1986).
Hattman, S. & Sun, W. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind Pmom promoter DNA. Nucleic Acids Res. 25, 4385–4388 (1997).
Swinton, D. et al. Purification and characterization of the unusual nucleotide, α-N-(9-β-d-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl Acad. Sci. USA 80, 7400–7404 (1983).
Sternberg, N. & Coulby, J. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc. Natl Acad. Sci. USA 87, 8070–8074 (1990).
Yarmolinski, M. B. & Sternberg, N. Bacteriophage P1. In The Bacteriophages Vol. 1 (ed. Calendar, R.) 782–791 (Plenum Press, New York, 1988).
Zweiger, G., Marczynski, G. & Shapiro, L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235, 472–485 (1994).
Marczynski, G. T. & Shapiro, L. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Genet. 56, 625–656 (2002). A comprehensive review on the Caulobacter cell cycle, including the roles of CcrM methylation.
Robertson, G. T. et al. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182, 3482–3489 (2000).
Reisenauer, A. & Shapiro, L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21, 4969–4977 (2002).
Vanyushin, B. F., Tkacheva, S. G. & Belozersky, A. N. Rare bases in animal DNA. Nature 225, 948–949 (1970).
Hattman, S. DNA-[adenine] methylation in lower eukaryotes. Biochemistry (Mosc.) 70, 550–558 (2005). A recent review on the presence of m6A in eukaryotes, the biological importance of which might have been overlooked for decades.
Rogers, S. D., Rogers, M. E., Saunders, G. & Holt, G. Isolation of mutants sensitive to 2-aminopurine and alkylating agents and evidence for the role of DNA methylation in Penicillium chrysogenum. Curr. Genet. 10, 557–560 (1986).
Hattman, S., Kenny, C., Berger, L. & Pratt, K. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 135, 1156–1157 (1978).
Gutiérrez, J. C., Callejas, S., Borniquel, S. & Martin-González, A. DNA methylation in ciliates: implications in differentiation processes. Int. Microbiol. 3, 139–46 (2000).
van Etten, J. L. et al. DNA methylation of viruses infecting a eukaryotic Chlorella-like green alga. Nucleic Acids Res. 13, 3471–3478 (1985).
Zhang, Y., Nelson, M., Nietfeldt, J. W., Burbank, D. E. & Van Etten, J. L. Characterization of Chlorella virus PBCV-1 CviAII restriction and modification system. Nucleic Acids Res. 20, 5351–5356 (1992).
Gogarten, J. P. & Townsend, J. P. Horizontal gene transfer, genome innovation and evolution. Nature Rev. Microbiol. 3, 679–687 (2005).
Lutsenko, E. & Bhagwat, A. S. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support, and implications. Mutat. Res. 437, 11–20 (1999).
Antonarakis, S. E., Krawczak, M. & Cooper, D. N. Disease-causing mutations in the human genome. Eur. J. Pedriatr. 159, 173–178 (2000).
Poole, A., Penny, D. & Sjoberg, B. M. Confounded cytosine! Tinkering and the evolution of DNA. Nature Rev. Mol. Cell Biol. 2, 147–51 (2001).
Fedoreyeva, L. I. & Vanyushin, B. F. N6-adenine DNA-methyltransferase in wheat seedlings. FEBS Lett. 514, 305–308 (2002).
Acknowledgements
Work in our laboratories is supported by grants from the Lejeune Foundation (to D.W.), and from the Spanish Ministry of Education and the European Regional Fund (to J.C.). We are grateful to D. Low and F. Antequera for helpful discussions. D.W. thanks A. L. Benabid and F. Berger for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Restriction–modification (R–M) system
-
Bacterial mechanism of defence against invasion by foreign DNA (for example, viruses). They are composed of genes that encode a restriction enzyme and a modification methylase.
- Distributive enzyme
-
Enzyme that dissociates from its substrate after one round of catalysis.
- Processive enzyme
-
Enzyme that performs multiple cycles of catalysis without dissociating from its substrate.
- Transition mutation
-
A nucleotide substitution that changes a purine to a purine (A↔G) or a pyrimidine to a pymiridine (C↔T).
- Leucine-responsive regulatory protein
-
Global regulator of the bacterial cell that regulates gene expression in response to exogenous leucine and other metabolic signals.
- Adhesin
-
Bacterial surface protein that facilitates adhesion to host tissues.
- Redox-sensitive regulator
-
Protein that can exist in two different states in response to the redox potential of the cell.
- Conjugal transfer
-
Transfer of bacterial DNA on cell-to-cell contact.
- F sex factor
-
Plasmid that is present in certain Escherichia coli strains that can transfer chromosomal genes, which led to the discovery of bacterial conjugation.
- Translocase
-
A protein involved in the translocation of proteins across membranes, and in the integration of proteins into the cytoplasmic membrane.
- Salmonella pathogenicity island I
-
(SPI-1). Gene cluster of ∼40 kb, located on centisome 63 in the Salmonella chromosome. The products of SPI-1 are necessary for invasion of epithelial cells.
- M (microfold) cell
-
Cell type located in the Peyer's patches of the small intestine. M cells are involved in antigen transport and interact with Salmonella and other bacterial pathogens.
- Lysogen
-
Bacterial cell that carries a viral genome in a non-infectious, repressed state.
- Rolling-circle replication
-
Mode of DNA replication that uses a circular DNA molecule as a template to produce concatemers of linear DNA molecules.
- Headful mechanism
-
Introduction of DNA into a bacteriophage capsid in such a way that the length of the packaged DNA molecule is determined by the size of the capsid.
- Theta replication
-
Mode of DNA replication that uses a circular DNA molecule as a template to produce two circular DNA molecules.
- Genomic imprinting
-
Epigenetic mechanism in diploid organisms by which only one allele (maternal or paternal) is expressed.
- X-chromosome inactivation
-
Epigenetic silencing of most genes in one of the two X chromosomes in somatic cells of mammalian females.
Rights and permissions
About this article
Cite this article
Wion, D., Casadesús, J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol 4, 183–192 (2006). https://doi.org/10.1038/nrmicro1350
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1350
This article is cited by
-
6mA-METL-9 axis regulates innate immunity in C. elegans
Cell Research (2023)
-
Genome-wide deposition of 6-methyladenine in human DNA reduces the viability of HEK293 cells and directly influences gene expression
Communications Biology (2023)
-
Bioprospection of the bacterial β-myrcene-biotransforming trait in the rhizosphere
Applied Microbiology and Biotechnology (2023)
-
Making a 6mA demethylase
Nature Chemical Biology (2022)
-
Advanced biotechnology using methyltransferase and its applications in bacteria: a mini review
Biotechnology Letters (2022)