Abstract
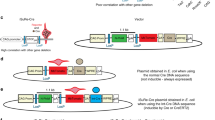
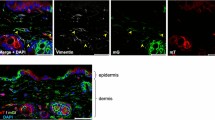
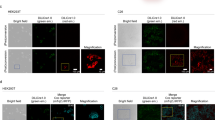
We developed a cell division–activated Cre-lox system for stochastic recombination of loxP-flanked loci in mice. Cre activation by frameshift reversion is modulated by DNA mismatch-repair status and occurs in individual cells surrounded by normal tissue, mimicking spontaneous cancer-causing mutations. This system should be particularly useful for delineating pathways of neoplasia, and determining the developmental and aging consequences of specific gene alterations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jonkers, J. & Berns, A. Nat. Rev. Cancer 2, 251–265 (2002).
Johnson, L. et al. Nature 410, 1111–1116 (2001).
Wang, W., Warren, M. & Bradley, A. Proc. Natl. Acad. Sci. USA 104, 4501–4505 (2007).
Lewandoski, M. & Martin, G.R. Nat. Genet. 17, 223–225 (1997).
Baker, S.M. et al. Cell 82, 309–319 (1995).
Narayanan, L. et al. Proc. Natl. Acad. Sci. USA 94, 3122–3127 (1997).
Harfe, B.D. & Jinks-Robertson, S. Annu. Rev. Genet. 34, 359–399 (2000).
Soriano, P. Nat. Genet. 21, 70–71 (1999).
Wong, M.H., Hermiston, M.L., Syder, A.J. & Gordon, J.I. Proc. Natl. Acad. Sci. USA 93, 9588–9593 (1996).
Marshman, E., Booth, C. & Potten, C.S. Bioessays 24, 91–98 (2002).
Jackson, E.L. et al. Genes Dev. 15, 3243–3248 (2001).
Calabrese, P., Tavare, S. & Shibata, D. Am. J. Pathol. 164, 1337–1346 (2004).
McDonald, S.A. et al. Cell Cycle 5, 808–811 (2006).
Robine, S. et al. Proc. Natl. Acad. Sci. USA 82, 8488–8492 (1985).
Acknowledgements
We thank M. Wong, N. Erdeniz, J. Johnson, O. Reilly and K. MacDonald for critically reading the manuscript. This work was supported by US National Institutes of Health grants R37 GM32741 to R.M.L. and 2 RO1 CA 80077 to D.S.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Methods (PDF 96 kb)
Rights and permissions
About this article
Cite this article
Miller, A., Dudley, S., Tsao, JL. et al. Tractable Cre-lox system for stochastic alteration of genes in mice. Nat Methods 5, 227–229 (2008). https://doi.org/10.1038/nmeth.1183
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1183
This article is cited by
-
Genetically-directed Sparse Neuronal Labeling in BAC Transgenic Mice through Mononucleotide Repeat Frameshift
Scientific Reports (2017)
-
Ssp DnaE split-intein mediated split-Cre reconstitution in tobacco
Plant Cell, Tissue and Organ Culture (PCTOC) (2013)
-
Different phenotypic consequences of simultaneous versus stepwise Apc loss
Oncogene (2012)
-
Harnessing mismatch repair to model sporadic cancers
Nature Methods (2008)