Abstract
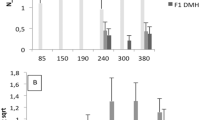
To dissect the multigenic control of colon tumour susceptibility in the mouse1 we used the set of 20 CcS/Dem (CcS) recombinant congenic (RC) strains2. Each CcS strain carries a unique, random subset of approximately 12.5% of the genome of strain STS/A (STS) on the genetic background of BALB/cHeA (BALB/c)3. Previously, applying a protocol of 26 injections of 1,2-dimethylhydrazine (DMH), we detected two susceptibility loci, Scc1 and Scc2, on chromosome 2 (refs 4, 5). Using a shorter tumour-induction procedure, combining DMH and N-ethyl-N-nitrosourea (ENU) treatment, we demonstrate that BALB/c, STS and most CcS strains are relatively resistant. The strain CcS-19, however, is susceptible, probably due to a combination of BALB/c and STS alleles at several loci. Analysis of 192 (BALB/c × CcS-19) F2 mice revealed, in addition to the Scc1/Scc2 region, three new susceptibility loci: Scc3 on chromosome 1, Scc4 on chromosome 17 and Scc5 on chromosome 18. Scc4 and Scc5 have no apparent individual effect, but show a strong reciprocal interaction. Their BALB/c and STS alleles are not a priori susceptible or resistant but the genotype at one locus determines the effect of the allele at the second locus and vice versa. These findings and the accompanying paper on lung tumour susceptibility6 show that interlocus interactions are likely to be an important component of tumour susceptibility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Demant, P., Resolution of susceptibility to cancer — new perspectives. Sem. Cancer Biol. 3, 159–166 (1992).
Demant, P. & Hart, A.A.M. Recombinant congenic strains — a new tool for analyzing genetic traits determined by more than one gene. Immunogenet. 24, 416–422 (1986).
Moen, C.J.A. et al The recombinant congenic strains — a novel genetic tool applied to the study of colon tumor development in the mouse. Mamm. Genome 1, 217–227 (1991).
Moen, C.J.A., Snoek, M., Hart, A.A.M. & Demant, P. Scc1, a novel colon cancer susceptibility gene in the mouse: linkage to CD44 (Ly-24, Pgp1) on chromosome 2. Oncogene 7, 563–566 (1992).
Moen, C.J.A., Groot, P.C., Hart, A.A.M., Snoek, M. & Demant, P.w Fine mapping of colon tumor susceptibility (Scc) genes in the mouse, different from the genes known to be somatically mutated in colon cancer. Proc. Natl. Acad. Sci. USA 93, 1082–1086 (1996).
Fijneman, R.J.A., de Vries, S.S., Jansen, R.C. & Demant, P. Complex interactions of new quantitative trait loci, Sluc1, Sluc2, Sluc3 and Sluc4, that influence susceptibility to lung cancer in the mouse. Nature Genet. 14, 465–467 (1996).
Deschner, E.E., Long, F.C. & Maskens, A.P. Relationship between dose, time, and tumor yield in mouse dimethylhydrazine-induced colon tumors. Cancer Lett. 8, 23–28 (1979).
Groot, P.C. et al. The recombinant congenic strains for analysis of multigenic trate: genetic composition. FASEB J. 6, 2826–2835 (1992).
Stassen, A.P.M., Groot, P.C., Eppig, J.T. & Demant, P. Genetic composition of the recombinant congenic strains. Mamm. Genome 7, 55–58 (1996).
Dietrich, W.F. et al Genetic identification of Mom1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75, 631–639 (1993).
Fijneman, R.J.A. & Demant, P. A gene for susceptibility to small intestinal cancer, ssic1, maps to the distal part of mouse chromosome 4.Cancer Res. 55, 3179–3182 (1995).
MacPhee M. . et al The secretory phospholipase A2 gene is a candidate for the Mom1 locus, a major modifier of ApcMin-induced intestinal neoplasia. Cell 81, 957–966 (1995).
Wu S. et al. TGF-beta is an autocrine-negative growth regulator of human colon carcinoma FET cells in vivo as revealed by transfection of an antisense expression vector. J. Cell. Biol. 116, 187–196 (1992).
Glaser, K.B., Mobilio, B., Chang, J.Y. & Senko, N. Phospholipase A2 enzymes: regulation and inhibition. Trends Pharmacol. Sci. 14, 92–98 (1993).
Neuman, R.J. & Rice, J.P. Two-locus models of disease. Genet. Epidemiol. 9, 347–365 (1992).
Risch, N., Ghosh, S. & Todd, J.A. Statistical evaluation of multiple locus linkage data in experimental species and its relevance to human studies: the application to nonobese diabetic (NOD) mouse and human insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 53, 702–714 (1993).
Schork, N.J., Boehnke, M., Terwilliger, J.D. & Ott, J. Two-trart-locus linkage analysis: a powerful strategy for mapping complex genetic traits. Am. J. Hum. Genet. 53, 1127–1136 (1993).
Cheverud, J.M. & Routman, E.J. Epistasis and its contribution to genetic variance components. Genetics, 139, 1455–1461 (1995).
Lark, K.G., Chase, K., Adler, F., Mansur, L.M. & Orf, J.H. Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proc. Natl. Acad. Sci. USA 92, 4656–4660 (1995).
Coupland, G. Genetic and environmental control of flowering time in Arabidopsis. Trends Genet. 11, 393–397 (1995).
Richard, I. et al. Mutations in the proteolytic enzyme calpain 3 cause limb–girdle muscular dystrophy type 2A. Cell 81, 27–40 (1995).
Avner, P. Quantity and quality: polygenic analysis in the mouse. Nature Genet. 7, 3–4 (1994).
De Sanctis, G.T. et al. Quantitative locus analysis of airway hyperresponsiveness in A/J and C57BL/6J mice. Nature Genet. 11, 150–154 (1995).
Frankel, W.N. et al. New seizure frequency QTL and the complex genetics in EL mice. Mamm. Genome 6, 830–838 (1995).
Laird, P.W. et al. A simplified mammalian DNA isolation procedure. Nucl. Acid Res. 19, 4293 (1991).
Dietrich, W. et al. A genetic map for the mouse suitable for typing intraspecific crosses. Genetics 131, 423–447 (1992).
Lander, E.S. & Schork, N.J. Genetic dissection of complex traits. Science 265, 2037–2048 (1994).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wezel, T., Stassen, A., Moen, C. et al. Gene interaction and single gene effects in colon tumour susceptibility in mice. Nat Genet 14, 468–470 (1996). https://doi.org/10.1038/ng1296-468
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng1296-468
This article is cited by
-
Quantitative trait loci for resistance to Flavobacterium psychrophilum in rainbow trout: effect of the mode of infection and evidence of epistatic interactions
Genetics Selection Evolution (2018)
-
A novel locus on mouse chromosome 7 that influences survival after infection with tick-borne encephalitis virus
BMC Neuroscience (2018)
-
Mice with different susceptibility to tick-borne encephalitis virus infection show selective neutralizing antibody response and inflammatory reaction in the central nervous system
Journal of Neuroinflammation (2013)
-
Characterization of a major colon cancer susceptibility locus (Ccs3) on mouse chromosome 3
Oncogene (2010)
-
Linkage analysis of the genetic determinants of T-cell IL-4 secretion, and identification of Flj20274 as a putative candidate gene
Genes & Immunity (2005)