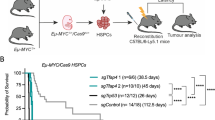
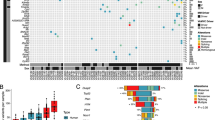
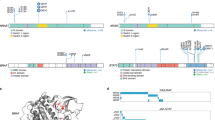
Abstract
We have screened the entire coding region of c–myc in a panel of Burkitt's lymphomas (BLs) and mouse plasmacytomas (PCTs). Contrary to the belief that c–myc is wild type in these tumours, we found that 65% of 57 BLs and 30% of 10 PCTs tested exhibit at least one amino acid (aa) substitution. These mutations were apparently homozygous in all BL cell lines tested and two tumour biopsies, implying that the mutations often occur before Myc/lg translocation in BL. In PCTs, only the mutant c–myc allele was expressed indicating a functional homozygosity, but occurrence of mutations after the translocation. Many of the observed mutations are clustered in regions associated with transcriptional activation and apoptosis, and in BLs, they frequently occur at sites of phosphorylation, suggesting that the mutations have a pathogenetic role.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Marcu, K.B., Bossone, S.A. & Patel, A.J. Myc function and regulation. Ann. Rev. Biochem. 61, 809–860 (1992).
Prendergast, G.C. & Ziff, E.B. A new bind for myc. Trends Genet. 8, 91–96 (1992).
Depinho, R.A., Schreiber-Agus, N. & Alt, F.W. Myc family oncogenes in the development of normal and neoplastic cells. Adv. cancer Res. 57, 1–46 (1991).
Cole, M. The myc oncogene: its role in transformation and differentiation. Adv. cancer Res. 20, 361–384 (1986).
Potter, M. & Wiener, F. Plasmacytomagenesis in mice: model; model of neoplastic development dependent upon chromosomal translocations. Carcinogenesis 13, 1681–1697 (1992).
Magrath, I. The pathogenesis of Burkitt's lymphoma. Adv. cancer Res. 55, 133–270 (1990).
Evan, G.I. et al. Induction of apoptosis in fibroblasts by c–Myc protein. Cell 69, 119–128 (1992).
Wyllie, A.H. et al. Rodent fibroblast tumours expressing human myc and ras genes: growth, metastasis and endogenous oncogene expression. Br. J. Cancer 56, 251–259 (1987).
Askew, D., Ashmun, R., Simmons, B. & Cleveland, J. Constitutive c–myc expression in IL–3 dependent myeloid cell line suppresses cycle arrest and accelerates apoptosis. Oncogene 6, 1915–1922 (1991).
Shi, Y. et al. Role for c–myc in activation-induced apoptotic cell death in T cell hybridomas. Science 257, 212–214 (1992).
Magrath, I., Jain, V. & Bhatia, K. Epstein-Barr virus and Burkitt's lymphoma. Sem. Cancer Biol. 3, 285–295 (1992).
Rabbitts, T.H., Hamlyn, R.H. & Baer, R. Altered nucleotide sequences of a translocated c–myc gene in Burkitt lymphoma. Nature 306, 760–765 (1983).
Rabbitts, T.H., Forster, A., Hamlyn, P. & Baer, R. Effect of somatic mutations within translocated c–myc genes in Burkitt lymphoma. Nature 309, 592–597 (1984).
Legouy, E. et al. Structure and expression of the murine L–myc gene. The EMBO J. 6, 3359–3366 (1987).
Gupta, S., Seth, A. & Davis, R.J. Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr–58 and Ser–62. Proc. natn. Acad. Sci. U.S.A. 90, 3216–3220 (1993).
Gu, W., Cechova, K., Tassi, V. & Dalla-Favera, R. Opposite regulation of gene transcription and cell proliferation by c–Myc and Max. Proc. natn. Acad. Sci. U.S.A. 90, 2935–2939 (1993).
Seth, A., Alvarez, E., Gupta, S. & Davis, R.J. A phosphorylation site located in the NH2-terminal domain of c–Myc increases transactivation of gene expression. J. biol. Chem. 266, 23521–23524 (1991).
Potter, M. et al. Avian v–myc replaces chromosomal translocation in murine plasmacytomagenesis. Science 235, 787–789 (1987).
Cory, S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv. cancer Res. 47, 189–234 (1986).
Suen, T.-C. & Hung, M.-C. & Hung, M.-C. c–myc reverses neu-induced transformed morphology by transcriptional repression. Molec. cell. Biol. 11, 354–362 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bhatia, K., Huppi, K., Spangler, G. et al. Point mutations in the c–Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet 5, 56–61 (1993). https://doi.org/10.1038/ng0993-56
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0993-56
This article is cited by
-
MYC directly transactivates CR2/CD21, the receptor of the Epstein–Barr virus, enhancing the viral infection of Burkitt lymphoma cells
Oncogene (2023)
-
Mistletoe extract Fraxini inhibits the proliferation of liver cancer by down-regulating c-Myc expression
Scientific Reports (2019)
-
Identification of MYC mutations in acute myeloid leukemias with NUP98–NSD1 translocations
Leukemia (2016)
-
A common functional consequence of tumor-derived mutations within c-MYC
Oncogene (2015)
-
Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis
Oncogene (2014)