Abstract
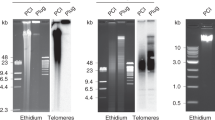
RecA–assisted restriction endonuclease (RARE) cleavage is a method to perform sequence–specific cleavage of genomic DNA, and is useful in physical mapping studies. After making two modifications, we have applied this method to mapping large regions of DNA in several cell types, including a notorious gap near the Huntington disease (HD) locus on chromosome 4. RARE cleavage fragments were analysed by pulsed field gel electrophoresis and Southern blotting and the distances between cleavage sites determined with accuracy. Using RARE cleavage, the gap measured was less than 60 kilobases in length. RARE cleavage is also a straightforward technique to map the distance from a marker to a telomere. The terminal 1.7 megabases of several HD and control cell lines were mapped with no large differences between cell lines in this region.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Ferrin, L.J. & Camerini-Otero, R.D. Selective cleavage of human DNA: RecA-Assisted Restriction Endonuclease (RARE) cleavage. Science 254, 1494–1497 (1991).
Ferrin, L.J. & Camerini-Otero, R.D. Synaptic complexes and RecA-assisted restriction endonuclease cleavage of DNA. Meth. molec. Genet. 2B (ed. Adolph, K.W.) 57–66 (Academic Press, Orlando, 1993).
Weber, B. et al. Genomlc organization and complete sequence of the human gene encoding the β-subunit of the cGMP phosphodiesterase and its localisation to 4p16.3. Nucl. Acids Res. 19, 6263–6268 (1991).
Whaley, W.L. et al. Mapping of cosmid clones in Huntington's disease region of chromosome 4. Somat. Cell molec. Genet. 17, 83–91 (1991).
Bates, G.P. et al. Defined physical limits of the Huntington's disease gene candidate region. Am. J. hum. Genet. 49, 7–16 (1991).
Bucan, M. et al. Physical maps of 4p16.3, the area expected to contain the Huntington disease mutation. Genomics 6, 1–15 (1990).
The Hungiton's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 (1993).
van den Engh, G., Sachs, R., & Trask, B.J. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science 257, 1410–1412 (1992).
Cox, D.R., Burmeister, M., Price, E.R., Kim, S. & Myers, R.M. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science 250, 245–250 (1990).
Bates, G.P. et al. A yeast artificial chromosome telomore clone spanning a possible location of the Huntington disease gene. Am. J. hum. Genet. 46, 762–775 (1990).
Gandelman, K., Gibson, L., Meyn, M.S., & Yang-Feng, T.L. Molecular definition of the smallest region of deletion overlap in the Wolf-Hirschhorn syndrome. Am. J. hum. Genet. 51, 571–578 (1992).
Wang, Y. & Schwartz, D.C., Chopped Inserts: a convenient alternative to agarose/DNA inserts or beads. Nucl. Acids Res. 21, 2528 (1993).
Altherr, M.R. et al. Radiation hybrid map spanning the Huntington disease gene region on chromosome 4. Genomics 13, 1040–1046 (1992).
Riethman, H.C., Spais, C., Buckingham, J., Grady, D. & Moyzis, R.K. Physical analysis of the terminal 240 kb of DNA from human chromosome 7q. Genomics 17, 25–32 (1993).
Foote, S., Vollrath, D., Hilton, A. & Page, D.C. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science 258, 60–66 (1992).
Chumakov, I. et al. Continuum of overlapping clones spanning the entire human chromosome 21q. Nature 359, 380–386 (1992).
Ichikawa, H. et al. A Notl restriction map of the entire long arm of human chromosome 21. Nature Genet. 4, 361–366 (1993).
Patterson, D. Mapping the way ahead. Nature Genet. 4, 323–324 (1993).
Schwartz, D.C., Li, X., Hernandez, L.I., Ramnarain, S.P., Huff, E.J. & Wang, Y. Ordered restriction maps of Saccnaromyces cerevisiae chromosomes constructed by optical mapping. Science 262,110–114 (1993).
McCombie, W.R. et al. Expressed genes, Alu repeats, and polymorphisms in cosmids sequenced from 4p16.3. Nature Genet. 1, 348–353 (1992).
Scott, H.S. et al. An 86-bp VNTR within IDUA is the basis of the D4S111 polymorphic locus. Genomics 14, 1118–1120 (1992).
Gusella, J.F. et al. Sequence-tagged sites (STSs) spanning 4p16.3 and the Huntington disease candidate region. Genomics 13, 75–80 (1992).
Feinberg, A.P. & Vogelstein, B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13 (1983) and 137, 266–267 (1984).
Herrmann, B.G. & Frischauf, A. Isolation of genomic DNA. Meth. Enzymol. 152 (eds Berger, S. L. & Kimmel, A.R.) 180–183 (Academic Press, Orlando, 1987).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ferrin, L., Camerini-Otero, R. Long–range mapping of gaps and telomeres with RecA–assisted restriction endonuclease (RARE) cleavage. Nat Genet 6, 379–383 (1994). https://doi.org/10.1038/ng0494-379
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0494-379
This article is cited by
-
A complete set of human telomeric probes and their clinical application
Nature Genetics (1996)