Abstract
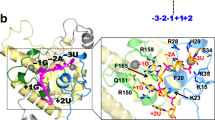
The Wilms' tumour suppressor gene 1 (WT1)1,2 encodes four C2H2 zinc finger-containing proteins3 critical for normal mammalian urogenital development4. Mutations in this gene are observed in the childhood kidney cancer, Wilms' tumour (WT)5. WT1 can bind specific DMA targets within the promoters of many genes6–9 and both transcriptional repression and activation domains have been identified10. On this basis, it has been assumed that regulation of transcription is the basis of WT1 tumour suppressor activity. However, subnuclear localization studies have revealed an association between WT1 proteins and ‘speckled bodies’ within the nucleus. Degradation of nuclear RNA in cells expressing WT1 abolishes this speckled localization and WT1 co-immunoprecipitates with a number of spliceosomal proteins, suggesting that it may also bind to RNA11. Using structural rather than sequence comparison, we have now identified an evolutionarily conserved N-terminal RNA recognition motif (RRM) in all known WT1 isoforms similar to that in the constitutive splicing factor U1A. Given the association between WT1 mutations and Wilms' tumours, this study, together with other recent findings, may suggest a novel tumour suppression mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Call, K.M. et al. Isolation and characterisation zinc finger polypeptide gene at the human chromosome 11 Wilms' tumour locus. Cell. 60, 509–520 (1990).
Gessler, M., Poustka, A., Cavenee, W., Neve, R.L., Orkin, S.H. & Bruns, G.A. Homozygous deletion in Wilms' tumours of a zinc-finger gene identified by chromosome jumping. Nature. 343, 774–778 (1990).
Haber, D.A., Sohn, R.L., Buckler, A.J., Pelletier, J., Cell, K.M. & Housman, D.E. Alternave splicing and genomic structure of the Wilms' tumor gene WT1. Proc. Natl. Acad. Sci. USA. 88, 9618–9622 (1991).
Kriedberg, J.A. et al. WT-1 is required for early kidney development. Cell. 74, 679–691 (1993).
Coppes, M.J., Campbell, C.E. & Williams, B.R.G. The role of WT1 in Wilms' tumorigenesis. FASEB J. 7, 886–895 (1993).
Rauscher, F.J. III, Morris, J.F., Tournay, O.E., Cook, D.M. & Curran, T. Binding of the Wilms' tumor locus zinc fingr protein to the EGR1 consensus sequence. Science. 250, 1259–1262 (1990).
Rausher, F.J. III. The WT1 Wilms' tumor gene product: a developmentally regulated transcription factor in the kidney that functions as a tumor suppressor. FASEB J. 7, 896–903 (1993).
Bickmore, W.A., Oghene, K., Little, M.H., Seawright, A., van Heyningen, V. & Hastie, N.D. Modulation of DMA binding specificity by alternative splicing of the Wilms' tumor WT1 gene transcript. Science. 257, 235–237 (1992).
Nakagama, H., Heinrich, G., Pelletier, J. & Housman, D.E. Sequence and structural requirements for high-affinity DMA binding by the WT1 gene product. Mol. Cell. Bio. 15, 1489–1498 (1995).
Wang, Z.-Y., Qui, Q.-Q. & Deuel, T.R. The Wilms' tumor gene product WT1 activates or suppresses transcription through separate functional domains. J. Biol. Chem. 268, 9172–9175 (1993).
Larsson, S.H. et al. Subnuclear localization of WT1 in splice-factor domains or transcription domains is regulated by alternative splicing. Cell 81, 391–401 (1995).
Smith, C.W.J., Patton, J.G. & Nadal-Ginard, B. Alternative splicing in the control of gene expression. Annu. Rev. Genet. 23, 527–577 (1989).
Kenan, D.J., Query, C.C. & Keene, J.D. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 16, 214–220 (1991).
Oubridge, C., Nobutoshi, I., Evans, P.R., Teo, C.H. & Nagai, K. Crystal structure at 1.92 Å resolution of the RNA-binding domian of the U1A spliceosomal portein complexed with an RNA hairpin. Nature. 372, 432–438 (1994).
Burd, C.G. & Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 265, 615–621 (1994).
Kim, Y.-J. & Baker, B.S. Isolation of RRM-type RNA-binding protein genes and the analysis of their relatedness by using a numerical approach. Mol. Cell. Biol. 13, 174–183 (1993).
Birney, E., Kurnar, S. & Krainer, A.R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucl. Acids Res. 21, 5803–6816 (1993).
Nagai, K., Oubridge, C., Jessen, T.H., Li, J. & Evans, P.R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 348, 515–520 (1990).
Kent, J., Coriat, A.M., Sharpe, P.T., Hastie, N. & Van Heyningen, V. The evolution of WT1 sequence and expression in the vertebrates. Oncogene. (in the press).
Joho, K., Darby, M.K., Crawford, E.T. & Brown, D.D. A finger protein structurally similar to TFIIIA that binds exclusively to 5S RNA in Xenopus. Cell. 61, 293–300 (1990).
Caricasole, A. et al. RNA binding by the Wilms tumor suppressor (WT1) zinc finger proteins. Proc. Natl. Acad. Sci. USA. (in the press).
Blundell, T.L., Sibanda, B.L., Sternberg, M.J.E. & Thornton, J.M. Knowledge-based prediction of protein structures and the design of novel molecules. Nature. 326, 347–362 (1987).
Chothia, C. et al. The predicted structure of immunoglobulin D1.3 and its comparison with the crystal structure. Science. 233, 755–758 (1986).
Amit, A.G., Mariuzza, R.A., Phillips, S.E.V. & Poljak, R.J. Three-dimensional structure of an antigen-antibody complex at 2.8(å) resolution. Science. 233, 747–753 (1986).
Driessen, H.P.C. & White, H. Molecular replacement and the crystallins. In Molecular Replacement. (ed. Machin, PA) 27–32, (Daresbury Laboratory, 1985).
Eisenberg, D., Wesson, M. & Yamashita, M. Interpretation of protein folding and binding with atomic solution parameters. Chem. Sci. 29A, 217–221 (1989).
Liithy, R., Bowie, J.U. & Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature. 356, 83–85 (1992).
Lüthy, R., McLachlan, A. & Eisenberg, D. Secondary structure-based profiles: use of structure-conserving scoring tables in searching protein sequence databases for structural similarities. Proteins. 10, 229–239 (1991).
Reddy, J.C. et al. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J. Biol. Chem. 270, 10878–10884 (1995).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kennedy, D., Ramsdale, T., Mattick, J. et al. An RNA recognition motif in Wilms' tumour protein (WT1) revealed by structural modelling. Nat Genet 12, 329–332 (1996). https://doi.org/10.1038/ng0396-329
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0396-329
This article is cited by
-
Sustained AWT1 expression by Dupuytren’s disease myofibroblasts promotes a proinflammatory milieu
Journal of Cell Communication and Signaling (2022)
-
WT1 expression is increased in primary fibroblasts derived from Dupuytren’s disease tissues
Journal of Cell Communication and Signaling (2015)
-
Localization of the Drosophila protein FL(2)D in somatic cells and female gonads
Cell and Tissue Research (2005)
-
Regulation of Gene Expression by WT1 in Development and Tumorigenesis
International Journal of Hematology (2002)
-
The possible role and application ofWT1 in Human Leukemia
International Journal of Hematology (2001)