Abstract
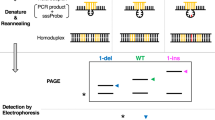
We have explored the application of the bacteriophage resolvases T4 endonuclease VII and T7 endonuclease I for detecting mutations in genomic DNA. Heteroduplex DNA fragments prepared by amplification from DNA containing known mutations were cleaved by one or both enzymes at nucleotide mismatches created by 3 of 3 short deletions and 13 of 14 point mutations in fragments as large as 940 basepairs. Heteroduplexes representing all four classes of possible single nucleotide mismatches were cleaved, and the sizes of the cleavage products generated correlated with the location of the mutation. We conclude that bacteriophage resolvases may be useful reagents for the rapid screening of DNA for mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Cotton, R.G. Current methods of mutation detection. Mutat Res. 285, 125–144 (1993).
Grompe, M. The rapid detection of unknown mutations in nucleic acids. Nature Genet. 5, 111–117 (1993).
Kemper, B. & Garabett, M. Studies on T4-head maturation. 1. Purification and characterization of gene-49-controlled endonuclease. Eur. J. Biochem. 115, 123–131 (1981).
Picksley, S.M., Parsons, C.A., Kemper, B. & West, S.C. Cleavage specificity of bacteriophageT4 endonuclease VII and bacteriophageT7 endonuclease I on synthetic branch migratable Holliday junctions. J. molec. Biol. 212, 723–735 (1990).
Myers, R.M., Maniatis, T. & Lerman, L.S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Meth. Enzym. 155, 501–527 (1987).
Orita, M., Suzuki, Y., Sekiya, T. & Hayashi, K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 4, 874–879 (1989).
White, M.B., Carvalho, M., Derse, D., O'Brien, S.J. & Dean, M. Detecting single base substitutions as heteroduplex polymorphisms. Genomics 5, 301–306 (1992).
Ganguly, A., Rock, M.J. & Prockop, D.J. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends In DNA heteroduplexes. Proc. natn. Acad. Sci. U.S.A. 90, 10325–10329 (1993).
Shenk, T.E., Rhodes, C., Rigby, P.W. & Berg, P. Mapping of mutational alterations In DNA with S1 nuclease: the location of deletions, Insertions and temperature-sensitive mutations in SV40. Cold Spring Harb. Symp. Quant. Biol. 1, 61–67 (1975).
Myers, R.M., Larin, Z. Maniatis, T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science 230, 1242–1246 (1985).
Lu, A.L. & Hsu, I.C. Detection of single DNA base mutations with mismatch repair enzymes. Genomics 14, 249–255 (1992).
Lishanski, A., Ostrander, E.A. & Rino, J. Mutation detection by mismatch binding protein, MutS, in amplified DNA: application to the cystic fibrosis gene. Proc. natn. Acad. Sc. U.S.A. 91, 2674–2678 (1994).
Ellis, L.A., Taylor, G.R., Banks, R. & Baumberg, S. MutS binding protects heteroduplex DNA from exonuclease digestion in vitro: a simple method for detecting mutations. Nucl. Acids Res. 22, 2710–2711 (1994).
Smooker, P.M. & Cotton, R.G. The use of chemical reagents in the detection of DNA mutations. Mutat. Res. 288, 65–77 (1993).
Ganguly, A. & Prockop, D.J. Detection of single-base mutations by reaction of DNA heteroduplexes with a water-soluble carbodlimldefollowed by primer extension: application to products from the polymerase chain reaction. Nucl. Acids Res. 18, 3933–3939 (1990).
Solaro, P.C., Blrkenkamp, K., Pfeiffer, P. & Kemper, B. Endonuclease VII of phage T4 triggers mismatch correction In vitro. J. molec. Blol. 230, 868–877 (1993).
Verpy, E., Biasotto, M., Meo, T. & Tosi, M. Efficient detection of point mutations on color-coded strands of target DNA. Proc. natn. Acad. Sci. U. S.A. 91, 1873–1877 (1994).
Villarejo, M.R. & Zabin, I. Beta-galactosldase from termination and deletion mutant strains. J. Bacteriol. 120, 466–474 (1974).
Kosak, H.G. & Kemper, B.W. Large-scale preparation of T4 endonuclease VII from over-expressing bacteria. Eur. J. Biochem. 194, 779–784 (1990).
Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K. & Mattick, J.S. Touchdown PCR to circumvent spurious priming during gene amplification. Nucl. Acids Res. 19, 4008 (1991).
Jiricny, J., Su, S.-S., Wood, S.G. & Modrich, P. Mismatch containing oligonucleotides bound by the E. coli mut-S-encoded protein. Nucl. Acids Res. 18, 7843–7853 (1988).
Davidow, L.S. Selecting PCR designed mismatch primers to create diagnostic restriction sites. Comp. Appl. Biosci. 8, 193–194 (1992).
Groden, J. et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66, 589–600 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Mashal, R., Koontz, J. & Sklar, J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet 9, 177–183 (1995). https://doi.org/10.1038/ng0295-177
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0295-177
This article is cited by
-
Noncanonical functions of Ku may underlie essentiality in human cells
Scientific Reports (2023)
-
Gene Editing in Human Haematopoietic Stem Cells for the Treatment of Primary Immunodeficiencies
Molecular Diagnosis & Therapy (2023)
-
A pair of primers facing at the double-strand break site enables to detect NHEJ-mediated indel mutations at a 1-bp resolution
Scientific Reports (2022)
-
One-Step Genotyping Method in loxP-Based Conditional Knockout Mice Generated by CRISPR-Cas9 Technology
Molecular Biotechnology (2022)
-
Multiplexed bioluminescence-mediated tracking of DNA double-strand break repairs in vitro and in vivo
Nature Protocols (2021)