Abstract
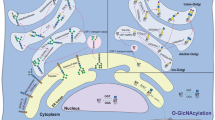
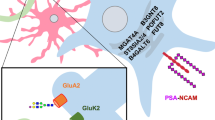
O-GlcNAc glycosylation is a unique, dynamic form of glycosylation found on intracellular proteins of all multicellular organisms. Studies suggest that O-GlcNAc represents a key regulatory modification in the brain, contributing to transcriptional regulation, neuronal communication and neurodegenerative disease. Recently, several new chemical tools have been developed to detect and study the modification, including chemoenzymatic tagging methods, quantitative proteomics strategies and small-molecule inhibitors of O-GlcNAc enzymes. Here we highlight some of the emerging roles for O-GlcNAc in the nervous system and describe how chemical tools have significantly advanced our understanding of the scope, functional significance and cellular dynamics of this modification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Torres, C.R. & Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 (1984).
Love, D.C. & Hanover, J.A. The hexosamine signaling pathway: deciphering the 'O-GlcNAc code'. Sci. STKE 2005, re13 (2005).
Roquemore, E.P., Chevrier, M.R., Cotter, R.J. & Hart, G.W. Dynamic O-GlcNAcylation of the small heat shock protein αB-crystallin. Biochemistry 35, 3578–3586 (1996).
Comer, F.I. & Hart, G.W. O-GlcNAc and the control of gene expression. Biochim. Biophys. Acta 1473, 161–171 (1999).
Zachara, N.E. & Hart, G.W. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 1673, 13–28 (2004).
Fulop, N., Marchase, R.B. & Chatham, J.C. Role of protein O-linked N-acetyl-glucosamine in mediating cell function and survival in the cardiovascular system. Cardiovasc. Res. 73, 288–297 (2007).
Wells, L. & Hart, G.W. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 546, 154–158 (2003).
Wells, L., Whelan, S.A. & Hart, G.W. O-GlcNAc: a regulatory post-translational modification. Biochem. Biophys. Res. Commun. 302, 435–441 (2003).
Slawson, C. & Hart, G.W. Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr. Opin. Struct. Biol. 13, 631–636 (2003).
Wells, L., Vosseller, K. & Hart, G.W. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 60, 222–228 (2003).
Lefebvre, T. et al. Does O-GlcNAc play a role in neurodegenerative diseases? Expert Rev. Proteomics 2, 265–275 (2005).
Lefebvre, T., Caillet-Boudin, M.L., Buee, L., Delacourte, A. & Michalski, J.C. O-GlcNAc glycosylation and neurological disorders. Adv. Exp. Med. Biol. 535, 189–202 (2003).
Kreppel, L.K., Blomberg, M.A. & Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 (1997).
Gao, Y., Wells, L., Comer, F.I., Parker, G.J. & Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 (2001).
Akimoto, Y. et al. Localization of the O-GlcNAc transferase and O-GlcNAc-modified proteins in rat cerebellar cortex. Brain Res. 966, 194–205 (2003).
Cole, R.N. & Hart, G.W. Cytosolic O-glycosylation is abundant in nerve terminals. J. Neurochem. 79, 1080–1089 (2001).
O'Donnell, N., Zachara, N.E., Hart, G.W. & Marth, J.D. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 (2004).
Lamarre-Vincent, N. & Hsieh-Wilson, L.C. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J. Am. Chem. Soc. 125, 6612–6613 (2003).
Khidekel, N. et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 (2007).
Griffith, L.S., Mathes, M. & Schmitz, B. β-Amyloid precursor protein is modified with O-linked N-acetylglucosamine. J. Neurosci. Res. 41, 270–278 (1995).
Griffith, L.S. & Schmitz, B. O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur. J. Biochem. 262, 824–831 (1999).
Lazarus, B.D., Love, D.C. & Hanover, J.A. Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421 (2006).
Kreppel, L.K. & Hart, G.W. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 274, 32015–32022 (1999).
Jinek, M. et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol. 11, 1001–1007 (2004).
Iyer, S.P., Akimoto, Y. & Hart, G.W. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J. Biol. Chem. 278, 5399–5409 (2003).
Brickley, K., Smith, M.J., Beck, M. & Stephenson, F.A. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J. Biol. Chem. 280, 14723–14732 (2005).
Stowers, R.S., Megeath, L.J., Gorska-Andrzejak, J., Meinertzhagen, I.A. & Schwarz, T.L. Axonal transport of mitochondria to synapses depends on Milton, a novel Drosophila protein. Neuron 36, 1063–1077 (2002).
Yang, X., Zhang, F. & Kudlow, J.E. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 (2002).
Wells, L., Kreppel, L.K., Comer, F.I., Wadzinski, B.E. & Hart, G.W. O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J. Biol. Chem. 279, 38466–38470 (2004).
Wells, L. et al. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic β-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 277, 1755–1761 (2002).
Toleman, C., Paterson, A.J., Whisenhunt, T.R. & Kudlow, J.E. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 279, 53665–53673 (2004).
Comtesse, N., Maldener, E. & Meese, E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a β-N-acetylglucosaminidase. Biochem. Biophys. Res. Commun. 283, 634–640 (2001).
Kelly, W.G. & Hart, G.W. Glycosylation of chromosomal proteins: localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell 57, 243–251 (1989).
Jackson, S.P. & Tjian, R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55, 125–133 (1988).
Zachara, N.E. & Hart, G.W. Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761, 599–617 (2006).
Yang, X. et al. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. USA 98, 6611–6616 (2001).
Roos, M.D., Su, K., Baker, J.R. & Kudlow, J.E. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 17, 6472–6480 (1997).
Gewinner, C. et al. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J. Biol. Chem. 279, 3563–3572 (2004).
Yang, W.H. et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 (2006).
Carlezon, W.A. Jr, Duman, R.S. & Nestler, E.J. The many faces of CREB. Trends Neurosci. 28, 436–445 (2005).
Mayr, B. & Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 (2001).
Levkovitz, Y. & Baraban, J.M. A dominant negative inhibitor of the Egr family of transcription regulatory factors suppresses cerebellar granule cell apoptosis by blocking c-Jun activation. J. Neurosci. 21, 5893–5901 (2001).
Jones, M.W. et al. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 4, 289–296 (2001).
Khidekel, N., Ficarro, S.B., Peters, E.C. & Hsieh-Wilson, L.C. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA 101, 13132–13137 (2004).
Wilson, M. & Koopman, P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 12, 441–446 (2002).
Rodda, D.J. et al. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 (2005).
Komitova, M. & Eriksson, P.S. Sox-2 is expressed by neural progenitors and astroglia in the adult rat brain. Neurosci. Lett. 369, 24–27 (2004).
Collart, M.A. Global control of gene expression in yeast by the Ccr4–Not complex. Gene 313, 1–16 (2003).
Vosseller, K. et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 (2006).
Wells, L. et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 (2002).
Deller, T. et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc. Natl. Acad. Sci. USA 100, 10494–10499 (2003).
Vazquez, L.E., Chen, H.J., Sokolova, I., Knuesel, I. & Kennedy, M.B. SynGAP regulates spine formation. J. Neurosci. 24, 8862–8872 (2004).
Sala, C. et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31, 115–130 (2001).
Israely, I. et al. Deletion of the neuron-specific protein δ-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 14, 1657–1663 (2004).
Murphy, J.E., Hanover, J.A., Froehlich, M., DuBois, G. & Keen, J.H. Clathrin assembly protein AP-3 is phosphorylated and glycosylated on the 50-kDa structural domain. J. Biol. Chem. 269, 21346–21352 (1994).
Yoshimura, T. et al. GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 120, 137–149 (2005).
Arnold, C.S. et al. The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J. Biol. Chem. 271, 28741–28744 (1996).
Dong, D.L., Xu, Z.S., Hart, G.W. & Cleveland, D.W. Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J. Biol. Chem. 271, 20845–20852 (1996).
Griffith, L.S. & Schmitz, B. O-linked N-acetylglucosamine is upregulated in Alzheimer brains. Biochem. Biophys. Res. Commun. 213, 424–431 (1995).
Liu, F., Iqbal, K., Grundke-Iqbal, I., Hart, G.W. & Gong, C.X. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. USA 101, 10804–10809 (2004).
Ludemann, N. et al. O-glycosylation of the tail domain of neurofilament protein M in human neurons and in spinal cord tissue of a rat model of amyotrophic lateral sclerosis (ALS). J. Biol. Chem. 280, 31648–31658 (2005).
Van Tine, B.A., Patterson, A.J. & Kudlow, J.E. Assignment of N-acetyl-D-glucosaminidase (Mgea5) to rat chromosome 1q5 by tyramide fluorescence in situ hybridization (T-FISH): synteny between rat, mouse and human with insulin degradation enzyme (IDE). Cytogenet. Genome Res. 103, 202B (2003).
Nolte, D. & Muller, U. Human O-GlcNAc transferase (OGT): genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm. Genome 13, 62–64 (2002).
Ballatore, C., Lee, V.M. & Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 (2007).
Robertson, L.A., Moya, K.L. & Breen, K.C. The potential role of tau protein O-glycosylation in Alzheimer's disease. J. Alzheimers Dis. 6, 489–495 (2004).
Lefebvre, T. et al. Evidence of a balance between phosphorylation and O-GlcNAc glycosylation of Tau proteins–a role in nuclear localization. Biochim. Biophys. Acta 1619, 167–176 (2003).
Alexander, G.E., Chen, K., Pietrini, P., Rapoport, S.I. & Reiman, E.M. Longitudinal PET evaluation of cerebral metabolic decline in dementia: a potential outcome measure in Alzheimer's disease treatment studies. Am. J. Psychiatry 159, 738–745 (2002).
Li, X., Lu, F., Wang, J.Z. & Gong, C.X. Concurrent alterations of O-GlcNAcylation and phosphorylation of tau in mouse brains during fasting. Eur. J. Neurosci. 23, 2078–2086 (2006).
Zhang, F. et al. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 (2003).
Liu, K. et al. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J. Neurochem. 89, 1044–1055 (2004).
Ciechanover, A. & Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40, 427–446 (2003).
Golks, A., Tran, T.T., Goetschy, J.F. & Guerini, D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 26, 4368–4379 (2007).
Liu, K., Paterson, A.J., Chin, E. & Kudlow, J.E. Glucose stimulates protein modification by O-linked GlcNAc in pancreatic β cells: linkage of O-linked GlcNAc to β cell death. Proc. Natl. Acad. Sci. USA 97, 2820–2825 (2000).
Rex-Mathes, M. et al. O-GlcNAc expression in developing and ageing mouse brain. Biochimie 83, 583–590 (2001).
Marshall, S., Bacote, V. & Traxinger, R.R. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 266, 4706–4712 (1991).
Haltiwanger, R.S., Blomberg, M.A. & Hart, G.W. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 267, 9005–9013 (1992).
Zachara, N.E. et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 (2004).
Greengard, P., Valtorta, F., Czernik, A.J. & Benfenati, F. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science 259, 780–785 (1993).
Soderling, T.R. CaM-kinases: modulators of synaptic plasticity. Curr. Opin. Neurobiol. 10, 375–380 (2000).
Colbran, R.J. Protein phosphatases and calcium/calmodulin-dependent protein kinase II-dependent synaptic plasticity. J. Neurosci. 24, 8404–8409 (2004).
Cieniewski-Bernard, C. et al. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 3, 577–585 (2004).
Roquemore, E.P., Chou, T.Y. & Hart, G.W. Detection of O-linked N-acetylglucosamine (O-GlcNAc) on cytoplasmic and nuclear proteins. Methods Enzymol. 230, 443–460 (1994).
Khidekel, N. et al. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem. Soc. 125, 16162–16163 (2003).
Ramakrishnan, B. & Qasba, P.K. Structure-based design of β1,4-galactosyltransferase I (β4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens β4Gal-T1 donor specificity. J. Biol. Chem. 277, 20833–20839 (2002).
Tai, H.C., Khidekel, N., Ficarro, S.B., Peters, E.C. & Hsieh-Wilson, L.C. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc. 126, 10500–10501 (2004).
Hsieh-Wilson, L.C., Khidekel, N., Arndt, S.E. & Tai, H.-C. Method and compositions for the detection of protein glycosylation. US patent application 20050130235 (2005).
Vocadlo, D.J., Hang, H.C., Kim, E.J., Hanover, J.A. & Bertozzi, C.R. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. USA 100, 9116–9121 (2003).
Wang, Z., Pandey, A. & Hart, G.W. Dynamic interplay between O-linked N-acetylglucos-aminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell. Proteomics 6, 1365–1379 (2007).
Syka, J.E., Coon, J.J., Schroeder, M.J., Shabanowitz, J. & Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 101, 9528–9533 (2004).
Nandi, A. et al. Global identification of O-GlcNAc-modified proteins. Anal. Chem. 78, 452–458 (2006).
Shafi, R. et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739 (2000).
Dauphinee, S.M., Ma, M. & Too, C.K. Role of O-linked β-N-acetylglucosamine modification in the subcellular distribution of α4 phosphoprotein and Sp1 in rat lymphoma cells. J. Cell. Biochem. 96, 579–588 (2005).
Forsythe, M.E. et al. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc. Natl. Acad. Sci. USA 103, 11952–11957 (2006).
Lee, T.N., Alborn, W.E., Knierman, M.D. & Konrad, R.J. Alloxan is an inhibitor of O-GlcNAc-selective N-acetyl-β-D-glucosaminidase. Biochem. Biophys. Res. Commun. 350, 1038–1043 (2006).
Meglasson, M.D., Burch, P.T., Berner, D.K., Najafi, H. & Matschinsky, F.M. Identification of glucokinase as an alloxan-sensitive glucose sensor of the pancreatic β-cell. Diabetes 35, 1163–1173 (1986).
Szkudelski, T. The mechanism of alloxan and streptozotocin action in B cells of the rat pancreas. Physiol. Res. 50, 537–546 (2001).
Gross, B.J., Kraybill, B.C. & Walker, S. Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 127, 14588–14589 (2005).
Dehennaut, V. et al. O-linked N-acetylglucosaminyltransferase inhibition prevents G2/M transition in Xenopus laevis oocytes. J. Biol. Chem. 282, 12527–12536 (2007).
Kim, E.J., Perreira, M., Thomas, C.J. & Hanover, J.A. An O-GlcNAcase-specific inhibitor and substrate engineered by the extension of the N-acetyl moiety. J. Am. Chem. Soc. 128, 4234–4235 (2006).
Kim, E.J. et al. Distinctive inhibition of O-GlcNAcase isoforms by an α-GlcNAc thiolsulfonate. J. Am. Chem. Soc. 129, 14854–14855 (2007).
Macauley, M.S., Whitworth, G.E., Debowski, A.W., Chin, D. & Vocadlo, D.J. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 280, 25313–25322 (2005).
Knapp, S. et al. Tautomeric modification of GlcNAc-thiazoline. Org. Lett. 9, 2321–2324 (2007).
Stubbs, K.A., Zhang, N. & Vocadlo, D.J. A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org. Biomol. Chem. 4, 839–845 (2006).
Dorfmueller, H.C. et al. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-GlcNAcylation levels. J. Am. Chem. Soc. 128, 16484–16485 (2006).
Shanmugasundaram, B. et al. Inhibition of O-GlcNAcase by a gluco-configured nagstatin and a PUGNAc-imidazole hybrid inhibitor. Chem. Commun. (Camb.) 13, 4372–4374 (2006).
Davis, B.G., Lloyd, R.C. & Jones, J.B. Controlled site-selective glycosylation of proteins by a combined site-directed mutagenesis and chemical modification approach. J. Org. Chem. 63, 9614–9615 (1998).
Carrillo, L.D., Krishnamoorthy, L. & Mahal, L.K. A cellular FRET-based sensor for β-O-GlcNAc, a dynamic carbohydrate modification involved in signaling. J. Am. Chem. Soc. 128, 14768–14769 (2006).
Ong, S.E., Mittler, G. & Mann, M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 (2004).
Zhang, Z. et al. A new strategy for the synthesis of glycoproteins. Science 303, 371–373 (2004).
Shin, Y. et al. Fmoc-based synthesis of peptide-thioesters: application to the total chemical synthesis of a glycoprotein by native chemical ligation. J. Am. Chem. Soc. 121, 11684–11689 (1999).
Schoof, H. et al. MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30, 91–93 (2002).
Acknowledgements
We thank C.J. Rogers and H.E. Murrey for critical reading of the manuscript. This work was supported in part by grants from the US National Institutes of Health (RO1 GM084724 and F31 NS056525-02 to J.E.R.) and the US National Science Foundation (CHE-0239861).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rexach, J., Clark, P. & Hsieh-Wilson, L. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol 4, 97–106 (2008). https://doi.org/10.1038/nchembio.68
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.68
This article is cited by
-
Glycometabolism change during Burkholderia pseudomallei infection in RAW264.7 cells by proteomic analysis
Scientific Reports (2022)
-
Loss of O-GlcNAcylation on MeCP2 at Threonine 203 Leads to Neurodevelopmental Disorders
Neuroscience Bulletin (2022)
-
O-GlcNAcylation: the “stress and nutrition receptor” in cell stress response
Cell Stress and Chaperones (2021)
-
Elevated O-GlcNAcylation induces an antidepressant-like phenotype and decreased inhibitory transmission in medial prefrontal cortex
Scientific Reports (2020)
-
Non-canonical amino acid labeling in proteomics and biotechnology
Journal of Biological Engineering (2019)