Abstract
Epidermal growth factor-like domain 7 (EGFL7) is a secreted factor implicated in cellular responses such as cell migration and blood vessel formation; however the molecular mechanisms underlying the effects of EGFL7 are largely unknown. Here we have identified transmembrane receptors of the Notch family as EGFL7-binding molecules. Secreted EGFL7 binds to a region in Notch involved in ligand-mediated receptor activation, thus acting as an antagonist of Notch signalling. Expression of EGFL7 in neural stem cells (NSCs) in vitro decreased Notch-specific signalling and consequently, reduced proliferation and self-renewal of NSCs. Such altered Notch signalling caused a shift in the differentiation pattern of cultured NSCs towards an excess of neurons and oligodendrocytes. We identified neurons as a source of EGFL7 in the brain, suggesting that brain-derived EGFL7 acts as an endogenous antagonist of Notch signalling that regulates proliferation and differentiation of subventricular zone-derived adult NSCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
02 July 2009
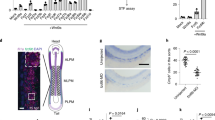
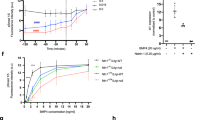
In the version of this article initially published online, the labelling of the yeast boxes in Fig. 1b was incorrect. In Fig. 1d the mNotch 2 values were missing, in Fig. 1g the Anti-EGFL7 was incorrectly labelled. In Fig 2c there was an extra ‘+’ sign. In Fig 3d the title was incorrect. In Fig 5b the BSA label was missing from under the fifth bar. These errors have been corrected in the HTML and PDF versions of the article.
References
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Selkoe, D. & Kopan, R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 26, 565–597 (2003).
Weinmaster, G. The ins and outs of notch signaling. Mol. Cell Neurosci. 9, 91–102 (1997).
Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M. E. Notch signaling. Science 268, 225–232 (1995).
Uyttendaele, H. et al. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 122, 2251–2259 (1996).
Weinmaster, G., Roberts, V. J. & Lemke, G. A homolog of Drosophila Notch expressed during mammalian development. Development 113, 199–205 (1991).
Rebay, I. et al. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67, 687–699 (1991).
Muskavitch, M. A. Delta-notch signaling and Drosophila cell fate choice. Dev. Biol. 166, 415–430 (1994).
Fleming, R. J., Scottgale, T. N., Diederich, R. J. & Artavanis-Tsakonas, S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 4, 2188–2201 (1990).
Ge, C., Liu, T., Hou, X. & Stanley, P. In vivo consequences of deleting EGF repeats 8–12 including the ligand binding domain of mouse Notch1. BMC Dev. Biol. 8, 48 (2008).
Tax, F. E., Yeargers, J. J. & Thomas, J. H. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368, 150–154 (1994).
Nichols, J. T., Miyamoto, A. & Weinmaster, G. Notch signaling—constantly on the move. Traffic 8, 959–969 (2007).
Mumm, J. S. & Kopan, R. Notch signaling: from the outside in. Dev. Biol. 228, 151–165 (2000).
Weinmaster, G. Notch signal transduction: a real rip and more. Curr. Opin. Genet. Dev. 10, 363–369 (2000).
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Honjo, T. The shortest path from the surface to the nucleus: RBP–J κ/Su(H) transcription factor. Genes Cells 1, 1–9 (1996).
Kageyama, R. & Ohtsuka, T. The Notch–Hes pathway in mammalian neural development. Cell Res. 9, 179–188 (1999).
Iso, T., Kedes, L. & Hamamori, Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 194, 237–255 (2003).
Bertrand, N., Castro, D. S. & Guillemot, F. Proneural genes and the specification of neural cell types. Nature Rev. Neurosci. 3, 517–530 (2002).
Soncin, F. et al. VE-statin, an endothelial repressor of smooth muscle cell migration. EMBO J. 22, 5700–5711 (2003).
Fitch, M. J., Campagnolo, L., Kuhnert, F. & Stuhlmann, H. Egfl7, a novel epidermal growth factor-domain gene expressed in endothelial cells. Dev. Dyn. 230, 316–324 (2004).
Parker, L. H. et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature 428, 754–758 (2004).
Carmeliet, P. & Tessier-Lavigne, M. Common mechanisms of nerve and blood vessel wiring. Nature 436, 193–200 (2005).
Strobl, L. J. et al. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP–Jκ. Immunobiology 198, 299–306 (1997).
Nyfeler, Y. et al. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. EMBO J. 24, 3504–3515 (2005).
Weng, A. P. et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell Biol. 23, 655–664 (2003).
Louvi, A. & Artavanis-Tsakonas, S. Notch signalling in vertebrate neural development. Nature Rev. Neurosci. 7, 93–102 (2006).
Yoon, K. & Gaiano, N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nature Neurosci. 8, 709–715 (2005).
Levy, O. A., Lah, J. J. & Levey, A. I. Notch signaling inhibits PC12 cell neurite outgrowth via RBP-J-dependent and -independent mechanisms. Dev. Neurosci. 24, 79–88 (2002).
Ohuchi, T., Maruoka, S., Sakudo, A. & Arai, T. Assay-based quantitative analysis of PC12 cell differentiation. J. Neurosci. Methods 118, 1–8 (2002).
Chitnis, A. Why is delta endocytosis required for effective activation of notch? Dev. Dyn. 235, 886–894 (2006).
Le Borgne, R., Bardin, A. & Schweisguth, F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132, 1751–1762 (2005).
Shimizu, K. et al. Integrity of intracellular domain of Notch ligand is indispensable for cleavage required for release of the Notch2 intracellular domain. EMBO J. 21, 294–302 (2002).
Hicks, C. et al. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J. Neurosci. Res. 68, 655–667 (2002).
Androutsellis-Theotokis, A. et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442, 823–826 (2006).
Morrison, S. J. et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101, 499–510 (2000).
Wang, S. et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron 21, 63–75 (1998).
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Bett, A. J., Haddara, W., Prevec, L. & Graham, F. L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl Acad. Sci. USA 91, 8802–8806 (1994).
Singec, I. et al. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nature Methods 3, 801–806 (2006).
Acknowledgements
We thank all our colleagues who provided constructs. Thanks to Julio Granados, Yonathan Lissanu Deribe and Koraljka Husnjak for initial help with the yeast two-hybrid system. Thanks to Verdon Taylor for helpful suggestions, Ruth von Laer for help with FACS analysis and Stefan Momma and David G. McEwan for comments on the manuscript. This work was financially supported by the DFG within the SFB Transregio 23, subproject A4 and the Excellence Cluster Cardiopulmonary System, by the Adolf Messer Foundation, by the Paul and Ursula Klein Foundation, and an EMBO long-term fellowship granted to M.H.H.S. (ALTF 2003-881).
Author information
Authors and Affiliations
Contributions
M.H.H.S. and I.D. designed experiments, analysed data and wrote the manuscript; I.D. supervised biochemical research; M.H.H.S. performed biochemical experiments and supervised neural and vascular experiments; F.B. worked on neurospheres and did IHC; I.N. and J.M. did experiments on D114 and HUVEC; T.B. did qRT-PCR; S.P. worked on protein purification; W.M.E. created EGFL7 antibodies and supported biochemical experiments; K.H.P. helped with experiments in brain and vasculature and all authors discsussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1751 kb)
Rights and permissions
About this article
Cite this article
Schmidt, M., Bicker, F., Nikolic, I. et al. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat Cell Biol 11, 873–880 (2009). https://doi.org/10.1038/ncb1896
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1896