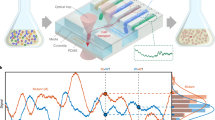
Abstract
We describe a genome-wide functional assay for rapid isolation of cell clones and genetic elements responsive to specific stimuli. A promoterless β-lactamase reporter gene was transfected into a human T-cell line to generate a living library of reporter-tagged clones. When loaded with a cell-permeable fluorogenic substrate, the cell library simultaneously reports the expression of a large number of endogenous genes. Flow cytometry was used to recover individual clones whose reporter-tagged genes were either induced or repressed following T-cell activation. Responsive clones were expanded and analyzed pharmacologically to identify patterns of regulation associated with specific genes. Although demonstrated using T cells, the genomic assay could be applied to map downstream transcriptional consequences for any propagating cell line in response to any stimulus of interest.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Heller, R.A., Schena, M., Chai, A., Shalon, D., Bedilion, T., Gilmore, J. et al. 1997. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc. Natl. Acad. Sci. USA 94: 2150– 2155.
Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. et al. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93: 10614–10619.
Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X.C., Stern, D. et al. 1996. Accessing genetic information with high-density arrays. Science 274: 610–614.
Skarnes, W.C., Auerbach, B.A., and Joyner, A.L. 1992. A gene trap approach in mouse embryonic stem cells: the lacZ reporter is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 6: 903–918.
Forrester, L.M., Nagy, A., Sam, M., Watt, A., Stevenson, L., Berstein, A. et al. 1996. An induction gene trap screen in embryonic stem cells: identification of genes that respond to retenoic acid in vitro. Proc. Natl. Acad. Sci. USA 93: 1677–1682.
Zlokarnik, G., Negulescu, P.A., Knapp, T.E., Mere, L., Burres, N., Feng., L. et al. 1998. Quantitation of transcription and clonal selection of single living cells using beta-lactamase as reporter. Science 279: 84–88.
Wurst, W., Rossant, J., Prideaux, V., Kownacka, M., Joyner, A., Hill, D.P. et al. 1995. A large-scale gene-trap screen for insertional mutation in developmentally regulated genes in mice. Genetics 139: 889–899.
Gogos, J.A., Lowry, W., and Karayiorgou, M. 1997. Selection for retroviral insertions into regulated genes. J. Virol. 71: 1644–1650.
Alberola-Ila, J., Takaki, S., Kerner, J.D., and Perlmutter, R.M. 1997. Differential signaling by lymphocyte antigen receptors. Annu. Rev. Immunol. 15: 125–154.
Truneh, A., Albert, F., Goldstein, P., and Schmitt-Verhulst, A.M. 1985. Early steps of lymphocyte activation bypassed by synergy between calcium ionophores and phorbol ester. Nature 313: 318–320.
Thastrup, O., Cullen, P.J., Drobak, B.K., Hanley, M.R., and Dawson, A.P. 1990. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. USA 87: 2466– 2470.
Takemura, H., Hughes, A.R., Thastrup, O., and Putney, J.W. 1989. Activation of calcium entry by the tumor promoter thapsigargin in parotid acinar cells. J. Biol. Chem. 264: 12266– 12271.
Rosenstreich, D.L. and Mizel, S.B. 1979 . Signal requirements for T lymphocyte activation: replacement of macrophage function with phorbol myristic acetate. J. Immunol. 123: 1749–1755.
Clipstone, N.A. and Crabtree, G.R. 1992 . Identification of calcineurin as a key signaling enzyme in T-lymphocyte activation. Nature 357: 695– 697.
Elliot, J.F., Lin, Y., Mizel, S.B., Bleackley, R.C., Harnish, D.G., and Paetkau, V. 1984. Induction of interleukin 2 messenger RNA inhibited by cyclosporin A. Science 226: 1439– 1443.
Negulescu, P.A., Shastri, N., and Calahan, M.D. 1994.Intracellular calcium dependence of gene expression in single T lymphocytes. Proc. Natl. Acad. Sci. USA 91: 2873–2877.
Fiering, S. and Al., E. 1990. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 4: 1823–1830.
Frohman, M.A., Dush, M.K., and Martin, G.R. 1988. Rapid production of full length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85: 8998 –9002.
Nagasawa, M., Melamed, I., Kupfer, A., Gelfand, E.W., and Lucas, J.J. 1997. Rapid nuclear translocation and increased activity of cyclin-dependent kinase 6 after T-cell activation. J. Immunol. 158: 5146–5154 .
Mages, H.W., Stamminger, T., Rilke, O., Bravo, R., and Kroczek, R.A. 1993. Expression of PILOT, a putative transcription factor, requires two signals and is cyclosporin A sensitive in T-cells. Int. Immunol. 5: 63–70.
Patwardhan, S., Gashler, A., Siegel, M.G., Chang, L.C., Joseph, L.J., Shows, T.B. et al. 1991. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene 6: 917–928.
Korchak, H.M., Kane, L.H., and Rossi, M.W. 1994. Long chain acyl coenzyme A and signaling in neutrophils. An inhibitor of acyl coenzyme A synthetase, triacsin C, inhibits superoxide anion generation and degranulation of human neutrophils. J. Biol. Chem. 269: 30281– 30287.
Faergeman, M.J. and Knudsen, J. 1997 . Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 323: 1–12.
Townley, D.J., Avery, B.J., Rosen, B., and Skarnes, W.C. 1997 .Rapid sequence analysis of gene trap integrations to generate a resourse of insertional mutations in mice. Genome Res. 7: 293–298.
Reddy, S., Rayburn, H., von Melcher, H., and Ruley, H.E. 1992. Fluorescence-activated sorting of titopotent embryonic stem cells expressing developmentally regulated lacZ fusion genes. Proc. Natl. Acad. Sci. USA 89: 6721– 6725.
Nolan, G.P., Fiering, S., Nicolas, J.F., and Herzenberg, L.A. 1988. Flourescence-activated cell analysis and sorting of viable mammalian cells based on β-D-galactosidase activity after transduction of E. coli lacZ. Proc. Natl. Acad. Sci. USA 85: 2603–2607.
Chowdury, H., Bonaldo, P., Torres, M., Stoykova, A., and Gruss, P. 1997. Evidence for the satochastic integration of gene trap vectors into the mouse germline. Nucleic Acids Res. q 25: 1531–1536.
Miller, S.A., Dykes, D.D., and Polesky, H.F. 1998. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215.
Acknowledgements
We thank Roger Tsien, Charles Zuker, Tom Curran, Kleanthis Xanthopoulos, and Timothy Rink for critical review of the manuscript. We also thank Bridgette Mandurrago for her technical assistance with DNA sequencing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Whitney, M., Rockenstein, E., Cantin, G. et al. A genome-wide functional assay of signal transduction in living mammalian cells. Nat Biotechnol 16, 1329–1333 (1998). https://doi.org/10.1038/4302
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/4302
This article is cited by
-
Determination of melatonin by a whole cell bioassay in fermented beverages
Scientific Reports (2019)
-
Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins
Nature Methods (2006)
-
Quantitative analysis of gene expression in living adult neural stem cells by gene trapping
Nature Methods (2005)
-
The art and design of genetic screens: mammalian culture cells
Nature Reviews Genetics (2004)
-
cAMP detection methods in HTS: selecting the best from the rest
Nature Reviews Drug Discovery (2004)