Abstract
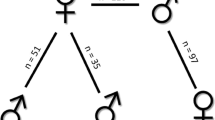
We report a unique application of the Random Amplified Polymorphic DNA (RAPD) technique for genetic linkage mapping of a single spruce tree using haploid DNA from megagametophyte tissue of individual seeds. Sixty–one segregating loci were analysed for reproducibility, inheritance and linkage. Forty–seven of the 61 markers were distributed into 12 linkage groups and covered 873.8 cM. The 14 markers not associated with any linkage group should be assigned to linkage groups as more markers are added to the map. This new approach quickly provides molecular genetic markers that are simple to evaluate for constructing genetic linkage maps and for other related genetic studies in forest tree species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Guries, R.P., Friedman, S.T. and Ledig, F.T. 1978. A megagameto phyte analysis of genetic linkage in pitch pine (Pinus rigida Mill.). Heredity 40: 309–314.
Rudin, D. and Ekberg, I. 1978. Linkage studies in Pinus sylvestris using macrogametophyte allozymes. Silvae genet 27: 1–12.
O'Malley, D.M., Allendorf, F.M. and Blake, G.M. 1979. Inheritance of isozyme variation and heterozygosity m Pinus ponderosa. Biochem Genet. 17: 233–250.
Eckert, R.T., Joly, R. and Neale, D.B. 1981. Genetics of isozyme variants and linkage relationships among allozyme loci in 35 eastern white pine clones. Can. J. For. Res. 11: 573–579.
Conkle, M.T. 1981. Isozyme variation and linkage in six conifer species, p. 11–17 In Proc. Sym. Isozymes of North American Forest Trees and Forest Insects. USDA For. Serv. Gen. Tech. Rep., PSW-48.
El-Kassaby, Y.A., Sziklai, O. and Yeh, F.C. 1982. Linkage relationships among 19 polymorphic allozyme loci in coastal Douglas-fir (Pseudotsuga menziesii var. ‘menziesii’). Can. J. Genet. Cytol. 124: 101–108.
Cheliak, W.M., Morgan, K., Dancik, B.P., Strobec, K.C. and Yeh, F.C. 1984. Segregation of allozymes in megagametophytes of viable seed from a natural population of jack pine, Pinus banksiana Lamb. Theor. Appl. Genet. 69: 145–151.
Strauss, S.H. and Conkle, M.T. 1986. Segregation, linkage, and diversity of allozymes in knobcone pine. Theor. Appl. Genet. 72: 483–493.
Helentjaris, R., Slocum, M., Schaeffer, A. and Nienhuis, J. 1986. Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor. Appl. Genet. 72: 761–769.
Helentjaris, H. 1987. A linkage map for maize based on RFLPs. TIG. 3: 217–221.
Burr, B. and Burr, F.A. 1991. Recombinant inbreds for molecular mapping in maize: theoretical and practical considerations. TIG 7: 55–60.
Beavis, W.D. and Grant, D. 1992. A linkage map based on information from F2 populations of maize (Zea mays L.). Theor. Appl. Genet. In press.
Bernatzky, R. and Tanksley, S. 1986. Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112: 887–898.
Keim, P., Shoemaker, R.C. and Palmer, R.G. 1989. Restriction fragment length polymorphism diversity in soybean. Theor. Appl. Genet. 77: 786–792.
McCouch, S.R., Kochert, G., Yu, Z.H., Wang, Z.Y., Kush, G.S., Coffman, W.R. and Tanksley, S.D. 1988. Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76: 815–829.
Harvey, M.J. and Muehlbauer, F.J. 1989. Linkages between restriction fragment length, isozyme and morphological markers in lentil. Theor. Appl. Genet. 77: 395–401.
Landry, B.S., Kesseli, R., Farrara, B. and Michelmore, R.W. 1987. A genetic map of lettuce (Lactuca sativa L. ) with restriction fragment length polymorphism, isozyme, disease resistance and morphological markers. Genetics 116: 331–337.
Gill, K.S., Lubbers, E.L., Gill, B.S., Raupp, W.J. and Cox, T.S. 1991. A genetic linkage map of Triticum tauschii (DD) and its relationship to the D genome of bread wheat (AABBDD). Genome 34: 362–372.
Heun, M., Kennedy, A.E., Anderson, J.A., Lapitan, N.L.V., Sorrells, M.E. and Tanksley, S.D. 1991. Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare). Genome 34: 437–444.
Slocum, M.K., Figdore, S.S., Kennard, W.C., Suzuki, Y.K. and Osborn, T.C. 1990. Linkage arrangement of restriction fragment length polymorphism loci in Brassica oleraceae. Theor. Appl. Genet. 80: 57–64.
Song, K.M., Suzuki, J.Y., Slocum, M.K., Williams, P.H. and Osborn, T.C. 1991. A linkage map of Brassica rapa (syn. campestns) based on restriction fragment length polymorphism loci. Theor. Appl. Genet. 82: 296–304.
Landry, B.S., Hubert, N., Etoh, T., Harada, J.J. and Lincoln, S.E. 1991. A genetic map for Brassica napus based on restriction fragment length polymorphisms detected with expressed DNA sequences. Genome 34: 543–552.
Chang, C., Bowman, A.W., Lander, E.S. and Meyerowitz, E.W. 1988. Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Proc. Natl. Acad. Sei. USA 85: 6856–6860.
Nam, H.G., Giraudat, J., den Boer, B., Moonan, F., Loos, W.D.B., Hauge, B.M. and Goodman, H.M. 1989. Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell 1: 699–705.
Williams, J.G.K., Kubelik, A.R., Livak, K.J., Rafalski, J.A. and Tingey, S.V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nuc. Acids Res. 18: 6531–6535.
Wagner, D.B., Furnier, G.R., Saghai-Maroof, M.A., Williams, S.M., Dancik, B.W., and Allard, R.W. 1987. Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc. Natl. Acad. Sei. USA 84: 2097–2100.
Carlson, J.E., Tulsieram, L.K., Glaubitz, J.C., Luk, V.W.K., Kauffeldt, C. and Rutledge, R. 1991. Segregation of random amplified DNA markers in F1 progeny of conifers. Theor. Appl. Genet. 83: 194–200.
Murray, G. and Skeates, D. 1985. Variation in height of White Spruce Provenances after 10 and 20 years in five field tests, p. 82–89. In: Proc. of 29th North Eastern Tree Improvement Conference. M.E. Demeritt (Ed. ). Published by North Eastern Tree Improvement Conference, P. O. Box 640, Durban, NH.
Ernst, S.G., Hanover, J.W., Keathley, D.E. and Mao, I.L. 1988. Genetic variation and control of intraspecific crossability in Blue and Englemann Spruce. Silvae Genetica 37: 112–118.
Yeh, F.C., Khalil, M.A.K., El-Kassaby, Y.A. and Trust, D.C. 1986. Allozyme variation in Picea mariana from Newfoundland: genetic diversity, population structure, and analysis of variation. Can. J. For. Res. 16: 713–720.
Alden, J. and Loopstra, C. 1987. Genetic diversity and population structure of Picea glauca on an altitudinal gradient in interior Alaska. Can. J. For. Res. 17: 1519–1526.
Edwards, M.D., Stuber, C.W. and Wendel, J.F. 1987. Molecular marker facilitated investigations of quantitative trait loci in maize. I. Numbers, genomic distribution and type of gene action. Genetics 116: 113–125.
Tulsieram, L.K., Compton, W.A., Morris, R., Thomas-Compton, M. and Eskridge, K. 1992. Analysis of genetic recombination in maize populations using molecular markers. Theor. Appl. Genet. In press
Neale, D.B. and Williams, C.G. 1991. Restriction fragment length polymorphism mapping in conifers and applications to forest genetics and tree improvement. Can. J. For. Res. 21: 545–554.
Nienhuis, J., Helentjaris, T., Slocum, M., Ruggero, B. and Shaefer, A. 1987. Restriction fragment length polymorphism analysis of loci associated with insect resistance in tomato. Crop Sci. 27: 797–803.
Osborn, T.C., Alexander, D.C. and Fobes, J.F. Identification of restriction fragment length polymorphisms linked to genes controlling soluble solids content in tomato fruit. Theor. Appl. Genet. 73: 350–356.
Stuber, C.W., Edwards, M.D. and Wendel, J.F. 1987. Molecular marker-facilitated investigations of quantitative trait loci in maize. II. Factors influencing yield and its component traits. Crop Sci. 27: 639–648.
Martin, B., Nienhuis, J., King, D. and Shaefer, A. 1989. Restriction fragment length polymorphisms associated with water use efficiency in tomato. Science 243: 1725–1728.
Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E. and Newberg, I. 1987. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181.
Suiter, K.A., Wendel, J.F. and Case, J.S. 1983. Linkage-1: a Pascal computer program for the detection and analysis of genetic linkage. J. Hered. 74: 203–204.
Kosambi, D.D. 1944. The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175.
Srivastava, P.S. and Johri, B.M. 1988. Pollen embryogenesis. J. Pathology 23–24: 83–99.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tulsieram, L., Glaubitz, J., Kiss, G. et al. Single Tree Genetic Linkage Mapping in Conifers Using Haploid DNA from Megagametophytes. Nat Biotechnol 10, 686–690 (1992). https://doi.org/10.1038/nbt0692-686
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0692-686
This article is cited by
-
High-throughput linkage mapping of Australian white cypress pine (Callitris glaucophylla) and map transferability to related species
Tree Genetics & Genomes (2015)
-
Narrow sense heritability estimates of bacterial leaf spot resistance in pseudo F2 (F1) population of mulberry (Morus spp.)
European Journal of Plant Pathology (2012)
-
RAPD Fingerprint to Appraise the Genetic Fidelity of In Vitro Propagated Araucaria excelsa R. Br. var. glauca Plantlets
Molecular Biotechnology (2012)
-
Genomics of a phylum distant from flowering plants: conifers
Tree Genetics & Genomes (2012)
-
Near-saturated and complete genetic linkage map of black spruce (Picea mariana)
BMC Genomics (2010)