Abstract
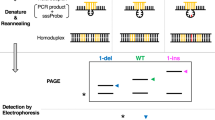
Flap endonucleases (FENs) isolated from archaea are shown to recognize and cleave a structure formed when two overlapping oligonucleotides hybridize to a target DNA strand. The downstream oligonucleotide probe is cleaved, and the precise site of cleavage is dependent on the amount of overlap with the upstream oligonucleotide. We have demonstrated that use of thermostable archaeal FENs allows the reaction to be performed at temperatures that promote probe turnover without the need for temperature cycling. The resulting amplification of the cleavage signal enables the detection of specific DNA targets at sub-attomole levels within complex mixtures. Moreover, we provide evidence that this cleavage is sufficiently specific to enable discrimination of single-base differences and can differentiate homozygotes from heterozygotes in single-copy genes in genomic DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lundquist, R.C. & Olivera, B.M. Transient generation of a displaced single stranded DNA during nick translation. Cell 31, 53–60 ( 1982).
Lyamichev, V., Brow, M.A.D. & Dahlberg, J.E. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science 260, 778–783 (1993).
Murante, R.S., Huang, L., Turchi, J.J. & Bambara, R.A. The calf 5´ to 3´ exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J. Biol. Chem. 269, 1191–1196 ( 1994).
Ceska, T.A., Sayers, J.R., Stier, G. & Suck, D. A helical arch allowing single-stranded DNA to thread through T5 5´-exonuclease. Nature 382, 90–93 ( 1996).
Lieber, M.R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19, 233–240 (1997).
Harrington, J.J. & Lieber, M.R. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 8, 1344–1355 (1994).
DeMott, M.S., Shen, B., Park, M.S., Bambara, R.A. & Zigman, S. Human RAD2 homolog 1 5´-to 3´-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5´ terminal abasic lesion by endonuclease activity. J. Biol. Chem. 271, 30068–30076 (1996).
O'Donovan, A., Davies, A.A., Moggs, J.G., West, S.C. & Wood, R.D. XPG endonuclease makes the 3´ incision in human DNA nucleotide excision repair. Nature 371, 432–435 (1994).
Bertina, R. Factor V Leiden and other coagulation factor mutations affecting thrombotic risk. Clin. Chem. 43, 1678– 1683 (1997).
Sferra, T.J. & Collins, F.S. The molecular biology of cystic fibrosis. Annu. Rev. Med. 44, 133– 144 (1993).
Lishanski, A., Ostrander, E.A. & Rine, J. Mutation detection by mismatch binding protein, MutS, in amplified DNA: application to the cystic fibrosis gene. Proc. Natl. Acad. Sci. USA 91, 2674–2678 (1994).
Copley, C.G. & Boot, C. Exonuclease cycling assay: an amplified assay for the detection of specific DNA sequences. Biotechniques 13, 888–892 ( 1992).
Okano, K. & Kambara, H. DNA probe assay based on Exonuclease III digestion of probes hybridized on target DNA. Anal. Biochem . 228, 101–108 ( 1995).
Duck, P., Alvarado-Urbina, G., Burdick, B. & Collier, B. Probe amplifier system based on chimeric cycling oligonucleotides. Biotechniques 9, 142–148 (1990).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular cloning: A laboratory manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989).
Acknowledgements
We thank Frank Robb for providing the sequence and the DNA containing the gene for the Pfu FEN1 nuclease, and we thank Laura Heisler, Lance Fors, Dennis Blakeslee, Lloyd Smith, and James Dahlberg for discussions and for critical reading of the manuscript. This work was supported by Cooperative Agreement 70NANB5H1030 from the Department of Commerce Advanced Technology Program to L. Fors, and by grant DE-FG02-94ER81891 from the Department of Energy to M. Brow.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lyamichev, V., Mast, A., Hall, J. et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat Biotechnol 17, 292–296 (1999). https://doi.org/10.1038/7044
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/7044
This article is cited by
-
Programmable microfluidic genotyping of plant DNA samples for marker-assisted selection
Microsystems & Nanoengineering (2018)
-
Survival analysis and pathological features of advanced non-small cell lung cancer with miliary pulmonary metastases in patients harboring epidermal growth factor receptor mutations
Journal of Cancer Research and Clinical Oncology (2018)
-
Clinical genetic strategies for early onset neurodegenerative diseases
Molecular & Cellular Toxicology (2018)