Abstract
The complexity of some sexual reproductive systems in arthropods still leaves both their genetic and epigenetic determinism and their evolutionary significance poorly understood. Pseudoarrhenotoky is characterized by obligate fertilization and differential inactivation and/or elimination of paternal chromosomes in embryos that develop into males. Here, we investigate how the paternal genome is transmitted in a pseudoarrhenotokous mite, Neoseiulus californicus, using codominant genetic markers detected by DALP (direct amplification of length polymorphism). Transmission patterns of parental alleles through one and two generations are reported at four or five loci corresponding to four linkage groups. Our data provide strong evidence for selective elimination of the paternal genome among male tissues. Sperm contained maternal genes exclusively, whereas some male somatic tissues retained most if not all paternal chromosomes. No recombination between parental genomes prior to paternal genome elimination from the embryonic germ line was observed. These data allow a reinterpretation of previous phenotypic and cytogenetic observations in these mites, from which we suggest some relevant mechanistic and evolutionary implications. In addition, this is the first published study using polymorphic codominant loci detected by the recently developed DALP method.
Similar content being viewed by others
Introduction
Early cytogenetic studies revealed a variety of systems of genetic transmission departing from conventional diplo-diploidy in several sexual insect and mite species. These systems were characterized by ‘highly aberrant meiotic mechanisms… combined with unusual mitotic phenomena during the cleavage divisions in the embryo’ (White, 1973 p. 501). These systems share the differential inactivation and/or elimination of chromosomes during embryonic development and gametogenesis; both between soma and germ line and between sexes. In fact, such phenomena appear consistently limited to male embryogenesis and spermatogenesis in several species belonging to such distant taxonomic groups as coccids (Homoptera) (reviewed in Nur, 1980), the coffee berry borer Hypothenemus hampei (Coleoptera: Scolytidae) (Brun et al., 1995; Borsa & Kjellberg, 1996) and phytoseiid mites (Acari: Mesostigmata) (Nelson-Rees et al., 1980; Schulten, 1985). In these species, both females and males arise from fertilized eggs, but in eggs developing into males, one set of chromosomes is inactivated during embryonic development and eliminated from the germ line or from the spermatocytes. So far, the study of these haplodiploid-like systems has been mainly concerned with the inheritance of phenotypic markers and radiation-induced chromosomal damage, and with the cytogenetics of embryonic development and spermatogenesis. All these studies have confirmed the maternal origin of the functional haploid chromosome set and the paternal origin of the inactivated or eliminated set in males (see Nur (1980) for a review on coccids, Schulten (1985) on phytoseiid mites and Brun et al. (1995) on the coffee berry borer).
These studies have also shown a high intertaxon variability in the extent (number of paternal chromosomes and amount/type of tissues) and timing (embryogenesis, spermatogenesis) of paternal chromosome elimination. This is particularly well documented within the Neococcoidea (Nur, 1980). Unlike in coccids, evidence for partial or complete haploidization of males is still ambiguous in pseudoarrhenotokous predatory mites (Acari: Phytoseiidae). Karyotyping has suggested true haploidy (Wysoki & Swirski, 1968; Oliver, 1977) as did phenotypic marker inheritance (Helle et al., 1978; Schulten, 1985), whereas radiation damage experiments were more consistent with the presence of paternal chromosomes in males in early embryonic development (Helle et al., 1978; Hoy, 1979). This ambiguity has led to various interpretations on the fate and function of paternal chromosomes in pseudoarrhenotokous mites. These include complete elimination and no function (Helle et al., 1978), a function restricted to early embryonic male development and subsequent complete elimination (Johanowicz & Hoy, 1998) and paternal genome retention throughout male life as a genetically inert chromatin mass (Hoy, 1979; Schulten, 1985). These hypotheses may translate into different predictions about the evolutionary role of pseudoarrhenotoky, including the advantages of male haploidy or the benefits of initiating embryonic development as a diploid for DNA recombination repair and masking of deleterious mutations (Sabelis & Nagelkerke, 1993).
A previous study in the phytoseiid mite Typhlodromus pyri has suggested the paternal transmission of two RAPD markers to males (Perrot-Minnot & Navajas, 1995). Study of the inheritance pattern was however, limited to two diagnostic bands and one generation. The objectives of the present study in the phytoseiid mite Neoseiulus californicus were therefore: (i) to determine the genetic constitution of F1 males using several locus-specific markers; (ii) to establish the transmission pattern of maternally inherited and potentially of paternally inherited alleles from F1 adult males to their daughters; and (iii) to derive a more general interpretation for the fate of the paternal genome in males of pseudoarrhenotokous mites. N. californicus is suspected to be pseudoarrhenotokous, as are most species in the family Phytoseiidae (Schulten, 1985; Norton et al., 1993; Sabelis & Nagelkerke, 1993), given that fertilization is necessary to initiate oviposition. We screened the mite genome for hypervariable markers with Mendelian transmission through a recently designed method, the Direct Amplification of Length Polymorphisms (DALP) (Desmarais et al., 1998). First and second generation progenies produced in heterogametic crosses and backcrosses, respectively, were genotyped at four or five codominant loci by PCR assay. These new data on paternal genome inheritance allowed a re-interpretation of genetic, cytogenetic and phenotypic data on pseudoarrhenotoky in mites.
Materials and methods
Origin and maintenance of N. californicus isofemale lines
We used genotyped isofemale lines in order to maximize allelic polymorphism at several loci in the controlled parental crosses. Several isofemale lines from N. californicus females collected in three orchards in October 1997, two in Montpellier and its vicinity (southern France), the other near Athens (Greece). Forty inbred lines were maintained for 25 generations, by isolating one sib-mated female upon emergence at each generation (from F1 to F8) or one gravid female every 10 days (the approximate generation time) (from F9 to F25). Single females were isolated on a pinto-bean leaf infected with all stages of the spider mite Tetranychus urticae as prey. The leaf was deposited on a black plastic sheet surrounded with glue, with its petiole trapped underneath in water-soaked cotton. The whole arena was enclosed in a plastic box, with two holes for ventilation. Rearing and crosses were maintained at 25°C, 60–70% RH and 16L:8D.
Identification of codominant loci and genotypic analysis
DALP is a method designed to detect polymorphic loci in any species, and it involves three steps: the production of multibanded (multilocus) pattern with a pair of arbitrary primers (around 20 bases long), re-amplification and direct sequencing of isolated bands, and definition of mono-locus-specific primers for PCR analysis (Desmarais et al., 1998). The isolation of codominant loci following DALP in the mite N. californicus is detailed elsewhere (Perrot-Minnot et al. submitted). Five polymorphic loci were isolated for which specific primers were designed in the conserved flanking regions of the polymorphism. Protocols for DNA extraction from a single mite and locus-specific PCR are given in Navajas et al. (1998) and Perrot-Minnot et al. (submitted), respectively. Radiolabelled PCR products were size-separated by PAGE. Therefore, allelic polymorphism was defined by fragment size variation of a single base or more. The genotypes of the 46 isofemale lines at the five loci were determined using five pooled females per line. We grouped several females per line in order to increase the probability of detecting within-line polymorphism (despite 25 generations of inbreeding). Two to five alleles per locus were identified among the 46 isofemale lines ranging in length from 132 to 198 bp and in frequency from 0.07 to 0.87 (Perrot-Minnot et al. submitted). No amplification product from the prey T. urticae was obtained with any of the five primer sets. Starvation was therefore no longer necessary before storing the N. californicus individuals at −20°C. Genotypic analysis of F1 and F2 progeny was performed by locus-specific PCR on single males and single females. Two controls for contamination were run at each PCR reaction, one negative amplification control (PCR reaction without DNA), one negative extraction control (DNA extraction and amplification without mite). These controls were always negative. We also checked the sequence homology between allelic bands in F1 sons and daughters by direct sequencing of PCR products. Two bands per locus (one per sex) were recovered from the gel and re-amplified. These PCR products were then purified (Wizards PCR Prep kit; Promega) and sequenced (Thermosequenase kit; APBiotech) (Perrot-Minnot et al. submitted).
Crossing design for parental and grand-paternal transmission and for analysis of recombination rate between the five loci
Pairs of lines to be crossed were chosen so as to maximize the number of loci polymorphic for the two parents (four or five). Twelve lines out of the 46 contained more than one allele at between one and three loci, so were not used. Both reciprocal crosses were performed with 20 pairs of lines by mating a virgin emerging female to a virgin male. Age and reproductive status were controlled by isolating single mites at the nymphal stage. Each couple was isolated for 24 h (mating usually occurred within two hours and lasted from 4 to 7 h) before transferring the female to a rearing and oviposition arena, designed as a modified Munger cell (Overmeer, 1985). To avoid any mixing between the mother and her emerging daughters, the female was transferred to a new arena every three days. After nine days of oviposition, the mother was removed and stored at −20°C. The progeny were allowed to develop to adulthood in their birth arena, then sexed, counted and stored at −20°C for further analysis (or −80°C for long-term storage). To study grand-paternal transmission, second-generation progenies from 23 parental crosses were produced by crossing one F1 male per family to a virgin female from its maternal line. Crosses and offspring collection were performed as above. These progeny are referred to as BCM (backcrossed F1 male progeny).
Recombination rates between pairs of loci were estimated in meiosis of F1 females heterozygous at the five loci. In order to get at least 30 offspring from genetically identical F1 females, two sisters were backcrossed to the same male from either the maternal or the paternal line. We refer to their progenies as BCF males or females (backcrossed F1 female progeny). Recombination rates were analysed by comparing the number of recombinant and parental genotypes of BCF females to the numbers expected under independent segregation using a Chi-squared test. Since multiple tests were simultaneously performed, the significance level of a test at a given pair of loci was adjusted according to the Bonferroni method (Holm, 1979).
Considering the offspring as a binomial population inheriting from one of the two paternal alleles (if any F1 ‘heterozygous’ males were produced), the proportion of female offspring inheriting the grand-paternal allele from their father corresponds to the binomial parameter p. We used the binomial test to determine the upper 95% confidence limit (L2) for the parameter p at a single locus, given the sample size (n, the number of BCM females genotyped) and the actual number of females inheriting the grand-paternal allele (=X ) (Zarr, 1996). Where X equalled zero, we used the exact computation L2=1 − (α/2)1/n (Zarr, 1996). The same test was used to determine the upper confidence limit for the proportion of recombinants among F1 males.
Results
Mendelian inheritance and recombination rates
Twenty-three F1 families were analysed for inheritance patterns at four loci. Since the final volume of extracted DNA (40 μL) allowed 7–8 PCR assays per single-mite sample, we were able to analyse transmission pattern at all loci in each single controlled cross. The genotypes of 23 F1 females (one per family) and 70 BCF females (produced by backcrossed heterozygous F1 females) were determined to check whether alleles showed Mendelian transmission. All F1 females analysed from heterogametic parental crosses were heterozygous at the polymorphic loci, therefore confirming biparental inheritance of the alleles and their codominance (Fig. 1). A balanced sorting of alleles was evidenced in F1 female meiosis from the genotypic proportions of heterozygous and homozygous BCF females at each locus (Chi-squared test, P=0.06–0.53) (Table 1, Fig. 2). Therefore, allelic inheritance through the diploid maternal phase verified that segregation and assortment followed Mendelian rules.
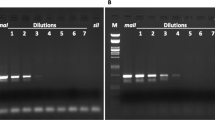
Autoradiogram showing the inheritance pattern of locus 235A in three families (a, b, c) produced in crosses between isofemale lines of the phytoseiid mite Neoseiulus californicus. M, maternal genotype; P, paternal genotype; S, F1 son; D, F1 daughter; (110)(100), alleles at locus 235A. Note complete homozygosity of F1 offspring in the homogametic cross (c).
Autoradiogram showing the inheritance pattern of locus 241B in a family of N. californicus produced by two heterozygous F1 sisters backcrossed to a male from their paternal line. M, maternal heterozygous genotype (100/120); P, paternal genotype; D, daughter; S, son. ‘Heterozygous’ males (100/[120]) typically exhibited weaker amplification pattern of the paternally inherited allele compared to the maternally inherited one, as observed at the previous generation at locus 235A (Fig. 1, F1 sons).
Recombination rates between the 10 pairs of loci were not significantly different from 0.5 after a Bonferroni adjustment, except for the pair 235A/234A (R=0.31) (Perrot-Minnot et al. submitted). A deficit in recombinants was found, suggesting a genetic distance of 31.43 ± 5.55 cM between these loci (GENE-LINK 2.0; Montagutelli, 1998). Therefore, we concluded that the five loci represented four linkage groups. Whether two groups could reside at opposite ends of the same chromosome or on distinct chromosomes cannot be established because of the lack of genetic map and more generally of cytogenetic data in mesostigmatid mites.
Paternal and maternal inheritance of the DALP alleles
A preliminary analysis was conducted on 23 F1 families to get an overview of transmission pattern of parental alleles to F1 males. The genotypes of two F1 males and one F1 female per family were determined at five loci. Because of low amplification yield and linkage to locus 235A, the locus 234A was not included in the F1 analysis. All 23 F1 females had a heterozygous genotype at either three or four loci as expected from the alleles fixed in the parental lines. Whenever the female was heterozygous, the electrophoretic pattern of her two brothers also showed two bands, one corresponding to the maternal allele and the other to the paternal allele (Fig. 1). The intensity of the latter band was consistently weaker than that of the maternally inherited allele, and weaker than the two parental bands in the corresponding sister (Fig. 1). Biparental transmission was therefore evident in males from heterogametic crosses but seemingly with a pattern different from that of true diploid sisters. This strong–weak inheritance pattern was consistent in all F1 males analysed for each locus and for each heterogametic cross (Table 1). Whenever homozygous parental genotypes were identical at a locus, both F1 males and F1 females were unambiguously homozygous (Fig. 1, Table 1).
Four families were further analysed to give a better estimate of within-family paternal transmission to F1 males. The genotypes of all F1 male offspring and both parents were determined at the four loci for each family. Because of a female-biased sex ratio, only 4–6 additional males could be analysed per family. The same pattern of biparental inheritance was evident in all F1 males at all polymorphic loci. The paternal allele was again consistently weaker than the maternal one (Fig. 1). Among the four families analysed, one stemmed from homogametic parents at locus 234C. In this case, a single band was found in all males corresponding to the common parental allele (Fig. 1, Table 1). In order to confirm the paternal origin of the weaker band in F1 males, eight bands were recovered from the gel and sequenced: two per locus, one from a male and one from its sister. In all cases, complete sequence homology was found between F1 male and female paternally inherited alleles, and between F1 bands and the original allelic sequence.
Since paternal genetic material was found in F1 males, we tested for its transmission to BCM females (produced in the backcross of F1 male to a female from his maternal line). Four groups of nine daughters and their parents were genotyped at five loci. None of these 36 grand-daughters was heterozygous, therefore showing that the grand-paternal allele detected in F1 males was not transmitted through the sperm (Fig. 3, Table 1). The 95% upper confidence limit for p, the proportion of offspring inheriting the grand-paternal allele from F1 male at a single locus, is 0.097.
Autoradiogram showing the inheritance pattern of locus 241B in two families of Neoseiulus californicus produced in backcrosses of F1 males to a female from their maternal line. (a) first family: M, maternal genotype (100/100); P, paternal genotype (100/[110]) showing the weakly paternally inherited allele (110); D, 10 daughters. (b) second family: M, maternal genotype (110/110); P, paternal genotype (110/[120]); D, 10 daughters. Note complete homozygosity of female offspring.
The relative amplification signals of maternally and paternally transmitted alleles in F1 males allowed the hypothesis of recombination between parental genomes preceding the partial elimination of the paternal genome to be tested. ‘Recombinant’ males would have had the reverse transmission pattern with the maternally inherited allele being less amplified than the paternal one. No ‘recombinant’ F1 males were recovered among the 43–59 adult males analysed. Given the minimum number of 43 males genotyped at any locus, the 95% upper confidence limit for p, the proportion of F1 ‘recombinant’ males, is 0.082. Furthermore, none of the 36 BCM females genotyped at four loci inherited from the grand-paternal allele, a transmission predicted if recombination occurred prior to paternal genome elimination from the germ line.
Discussion
The present study provides clear evidence for the retention of paternal genetic material in males of a pseudoarrhenotokous mite species. We expand on our previous report on paternal transmission of one RAPD marker in another species of phytoseiid mite (Perrot-Minnot & Navajas, 1995) in two important respects. First, we observed a consistent pattern of paternal transmission to males at four loci corresponding to four independent linkage groups. This is equivalent to the known diploid number of 8 chromosomes in N. californicus (Perrot-Minnot, unpubl. results) and in most species of Phytoseiidae (Wysoki & Swirski, 1968; Oliver, 1977; Wysoki & Bolland, 1983). We can thus confidently conclude that most if not all of the paternal genome is retained in males. However, the paternal genome is not transmitted through sperm, since paternal genes inherited by their daughters are exclusively of grand-maternal origin. Selective elimination of the paternal genome thus occurs in male tissues, resulting in sperm carrying only maternal genes. Haploidy of the male germ line thus categorizes pseudoarrhenotoky as a haplodiploid genetic system despite the diploid origin of males (White, 1973; Bull, 1979; Borgia, 1980).
Several hypotheses arise from this pattern of paternal chromosome inheritance in males relating to the extent of chromosome elimination and to the functional significance of somatic retention. Paternal chromosome elimination in male embryos of N. californicus may be germ-line limited, as in the lecanoid system of coccids and in sciarid flies (White, 1973; Nur, 1980), or could also affect some somatic tissues. Our observations of consistent differences in intensity between maternally inherited and paternally inherited alleles in F1 males support the latter hypothesis. This interpretation assumes that the lower amplification signal of paternal alleles resulted from a lower amount of paternal DNA retained in F1 males. Unequal amounts of target DNA are known to yield different amounts of amplified DNA as shown in quantitative PCR (Heid et al., 1996). This interpretation was supported by the lack of amplification of paternal alleles in some F1 male DNA extracts partially degraded by long-term storage and repeated freeze-thawing. Furthermore, most if not all of the paternal genome was retained in some somatic tissues based on the transmission of the alleles belonging to four linkage groups. This pattern of somatic retention of paternal chromosomes is unlikely to be stochastic, because it was identical in all F1 males analysed. Since spermatogenesis in phytoseiid mites is probably mitosis-like, as in Metaseiulus occidentalis (Nelson-Rees et al., 1980), the heterochromatic paternal chromosomes may be excluded early in embryonic development, at least from the germ line.
Two questions are of crucial interest to our understanding of such genetic systems, intermediate between diplo-diploidy and arrhenotoky: (i) the function, if any, of paternal genome retention; and (ii) the evolutionary significance of germ-line haploidization. These questions pertained to the evolution of haplodiploid-like systems (Bull, 1979; Borgia, 1980), with respect to the advantage of male haploidy and to the constraints of sex-determining mechanisms and haploid male development (either from unfertilized eggs or from eggs undergoing haploidization).
Four hypotheses on the function of paternal chromosomes have been put forward in pseudoarrhenotokous species of mites and coccids: (i) the paternal genome has no function and is eliminated during or soon after embryonic development; the increased male mortality and sterility following paternal genome damage result from a disruption of the regular process of haploidization (Helle et al., 1978); (ii) the integrity of the paternal genome is necessary to induce embryonic male development before its elimination (Johanowicz & Hoy, 1998); (iii) the paternal set is retained as a genetically inert chromatin mass throughout male life, fulfilling a residual function for dosage compensation (a ‘bulk’ function) (Nelson-Rees, 1962; Hoy, 1979; Schulten, 1985); and (iv) the paternal genes are expressed in some tissues following their reactivation (Nur, 1967, 1990). The first two hypotheses predict strict male haploidy and can therefore be rejected in at least two species of phytoseiid mites, N. californicus and T. pyri, based on the evidence for paternal gene retention in males (this study; Perrot-Minnot & Navajas (1995), respectively). Our data on paternal allele transmission to males cannot distinguish between the last two hypotheses, and further studies are needed to determine whether or not the paternal genome is genetically active. Most studies on male phenotypes were restricted to insecticide resistance involving the nervous system or gut and fat body in mites (several references in Schulten, 1985) and in the coffee berry borer (Brun et al., 1995; Borsa & Kjellberg, 1996), or to external morphology involving hypodermic cells (Nur, 1967). Despite showing exclusively maternal inheritance, the limited range of these characters may not have detected paternal gene expression in other tissues, as shown in coccids with a lecanoid system (Nur, 1967). In males of coccid species with a lecanoid system, paternal chromosomes are retained heterochromatized in both somatic and germinal tissues (from which they are excluded at the onset of spermatogenesis) (Nur, 1980). But reversal of the chromosomes of paternal origin to the euchromatic state has been observed in some somatic tissues (Nur, 1967). In particular, paternal gene expression or ‘functional diploidy’ of some tissues may be necessary for male fertility. To date, there is no evidence that a common mechanism controls both paternal genome inactivation/elimination and sex determination. Such a mechanism has been suggested by Brown & Chandra (1977) based on experimental evidence for maternal influences acting via the egg cytoplasm upon sex determination and paternal genome imprinting and heterochromatization.
The evolutionary advantages of pseudoarrhenotoky over arrhenotoky have been sought in the diploid origin of males and the retention of paternal genome in the early developmental period (Sabelis & Nagelkerke, 1993), such as the masking of deleterious mutations and recombination repair. Masking of deleterious maternal mutations during the period of paternal genome retention implies the early expression of paternal genes. This hypothesis has been dismissed in a coccid with a lecanoid system by Sabour (1972), showing that embryonic nuclear RNA was not synthesized earlier than the stage at which paternal genome heterochromatization occurred. Another evolutionary advantage, the recombination repair of the maternal genome via homologous sequences in the paternal genome, can also be questioned both theoretically and in relation to our data. Meiotic-like chromosome pairing was observed by Nelson-Rees et al. (1980) before reductional division and paternal genome elimination in 0–24 h-old embryos of the mite Metaseiulus occidentalis. The authors reported a diplotene-like pairing and opening out of three homologous chromosomes in embryos subsequently undergoing haploidization. It was thus suggested that recombination DNA repair of maternal chromosomes in males could occur (Sabelis & Nagelkerke, 1993). Our data, however, do not support the hypothesis of recombination between homologous duplexes following karyogamy and before paternal genome inactivation and eventual elimination from the germ line. In fact, the two ways of detecting recombination in F1, namely the relative intensity of amplicons between maternally and paternally inherited alleles in F1 males, and paternal alleles transmission to F2 females from ‘heterozygous’ males, did not revealed any. Thus, the hypothesis of recombination in males as an evolutionary advantage of pseudoarrhenotoky over arrhenotoky either in DNA repair or the generation of genetic variation can be rejected. Among the recognized evolutionary advantages of sexual reproduction in diploid organisms — DNA repair, enhanced genetic variation and complementation of gene function — (Bernstein et al., 1981), only the third one appears to be a possibility in pseudoarrhenotokous mites. However, complementation of gene function in male tissues retaining paternal genes remains questionable until the reversal of heterochromatization and paternal gene expression can be demonstrated, as in the lecanoid coccids (Nur, 1967, 1990). Further hypotheses for the evolution of pseudoarrhenotoky may involve theories of selfish DNA, genomic conflicts in the origin and maintenance of sex, or the importance of fertilization and developmental constraints. In particular, to clarify the relationship between selective elimination/retention of paternal chromosomes in males and sex determination may be quite decisive for our understanding of such constraints.
References
Bernstein, H., Byers, G. S. and Michod, R. E. (1981). Evolution of sexual reproduction: importance of DNA repair, complementation, and variation. Am Nat, 117: 537–549.
Borgia, G. (1980). Evolution of haplodiploidy: models for inbred and outbred systems. Theor Pop Biol, 17: 103–128.
Borsa, P. and Kjellberg, F. (1996). Experimental evidence for pseudo-arrhenotoky on Hypothenemus hampei (Coleoptera: Scolytidae). Heredity, 76: 130–135.
Brown, S. W. and Chandra, H. S. (1977). Chromosome imprinting and the differential regulation of homologous chromosomes. In: Goldstein, L. and Prescott, D. M. (eds) Cell Biology; A Comprehensive Treatise, vol. 1, pp. 109–189. Academic Press, New York.
Brun, L. O., Stuart, J., Gaudichon, V., Aronstein, K. and Ffrench-Constant, R. H. (1995). Functional haplodiploidy: a mechanism for the spread of insecticide resistance in an important international insect pest. Proc Natl Acad Sci USA, 92: 9861–9865.
Bull, J. J. (1979). An advantage for the evolution of male haploidy and systems with similar genetic transmission. Heredity, 43: 361–381.
Desmarais, E., Lanneluc, I. and Lagnel, J. (1998). Direct amplification of length polymorphism (DALP), or how to get and characterize new genetic markers in many species. Nucl Acids Res, 26: 1458–1465.
Heid, C. A., Stevens, J., Livak, K. J. and Williams, P. M. (1996). Real time quantitative PCR. Genome Res, 6: 986–994.
Helle, W., Bolland, H. R., van Arendonk, R., de Boer, R., Schulten, G. G. M. and Russell, V. M. (1978). Genetic evidence for biparental males in haplo-diploid predator mites (Acarina: Phytoseiidae). Genetica, 49: 165–171.
Holm, S. (1979). A simple sequentially rejective multiple test procedure. Scand J Stat, 6: 65–70.
Hoy, M. A. (1979). Parahaploidy of the ‘arrhenotokous’ predator Metaseiulus occidentalis (Acarina: Phytoseiidae) demonstrated by X-irradiation of males. Entomol Exp Appl, 26: 97–104.
Johanowicz, D. L. and Hoy, M. A. (1998). Experimental induction and termination of non-reciprocal reproductive incompatibilities in a parahaploid mite. Entomol Exp Appl, 87: 51–58.
Montagutelli, X. (1998). Gene-Link, version 2.0. Available at ftp: //ftp.pasteur.fr/pub/GenSoft/Ms-Windows/linkage_and_mapping/Gene-Link
Navajas, M., Lagnel, J., Gutierrez, J. and Boursot, P. (1998). Species-wide homogeneity of nuclear ribosomal ITS2 sequences in the spider mite Tetranychus urticae contrasts with extensive mitochondrial COI polymorphism. Heredity, 80: 742–752.
Nelson-Rees, W. A. (1962). The effect of radiation damaged heterochromatic chromosomes on male fertility in the mealy bug Planococcus citri (Risso). Genetics, 47: 661–683.
Nelson-Rees, W. A., Hoy, M. A. and Roush, R. T. (1980). Heterochromatinization, chromatin elimination and haploidization in the parahaploid mite Metaseiulus occidentalis (Nesbitt) (Acarina: Phytoseiidae). Chromosoma, 77: 263–276.
Norton, R. A., Kethley, J. B., Johnston, D. E. and O’connor, B. M. (1993). Phylogenetic perspectives on genetic systems and reproductive modes of mites. In: Wrensch, D. L. and Ebbert, M. A. (eds) Evolution and Diversity of Sex Ratio, pp. 8–99. Chapman & Hall, New York.
Nur, U. (1967). Reversal of heterochromatisation and the activity of the paternal chromosome set in the male mealy bug. Genetics, 56: 375–389.
Nur, U. (1980). Evolution of unusual chromosome systems in scale insects (Coccoidea: Homoptera). In: Blackman, R. L., Hewitt, G. M. and Ashburner, M. (eds) Insect Cytogenetics. Symposia of the Royal Entomological Society of London, no. 10, pp. 97–117. Blackwell Scientific Publications, Oxford.
Nur, U. (1990). Heterochromatisation and euchromatization of whole genomes in scale insects (Coccoidea: Homoptera). Development, (suppl.), 29–34.
Oliver, J. H. (1977). Cytogenetics of mites and ticks. Ann Rev Entomol, 22: 407–429.
Overmeer, W. P. J. (1985). Rearing and handling. In: Helle, W. and Sabelis, M. W. (eds) Spider Mites, Their Biology, Natural Enemies and Control. World Crop Pests, vol. 1B, pp. 161–170. Elsevier, Amsterdam.
Perrot-Minnot, M. -J. and Navajas, M. (1995). Biparental inheritance of RAPD markers in males of the pseudoarrhenotokous mite Typhlodromus pyri. Genome, 38: 838–844.
Sabelis, M. W. and Nagelkerke, C. J. (1993). Sex allocation and pseudoarrhenotoky in phytoseiid mites. In: Wrensch, D. L. and Ebbert, M. A. (eds) Evolution and Diversity of Sex Ratio, pp. 512–541. Chapman & Hall, New York.
Sabour, M. (1972). RNA synthesis and heterochromatisation in early development of a mealybug. Genetics, 70: 291–298.
Schulten, G. G. M. (1985). Pseudoarrhenotoky. In: Helle, W. and Sabelis, M. W. (eds) Spider Mites, Their Biology, Natural Enemies and Control. World Crop Pests, vol. 1B, pp. 67–71. Elsevier, Amsterdam.
White, M. J. D. (1973). Animal Cytology and Evolution, 3rd edn. Cambridge University Press, Cambridge.
Wysoki, M. and Bolland, H. R. (1983). Chromosome studies of phytoseiid mites (Acari: Gamasida). Int J Acarol, 9: 91–94.
Wysoki, M. and Swirski, E. (1968). Karyotypes and sex determination of ten species of phytoseiid mites (Acarina: Mesostigmata). Genetica, 39: 220–228.
Zarr, J. H. (1996). Biostatistical Analysis, 2nd edn. Prentice Hall International Editions, Upper Saddle River, N.J.
Acknowledgements
We thank Nicole Pasteur (Institut des Sciences de l’Evolution ISEM, laboratoire Génétique et Environnement, UMR-CNRS 5554, Université Montpellier II) for the use of laboratory facilities. Eric Desmarais, François Rousset and an anonymous reviewer are acknowledged for their useful comments on the manuscript. We thank Brigitte Cheval, Christophe Sauce and Marie-Sophie Simon for technical assistance, and Anastasia Tsagkarakou for supplying the Greek population of N. californicus.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Perrot-Minnot, MJ., Lagnel, J., Migeon, A. et al. Tracking paternal genes with DALP markers in a pseudoarrhenotokous reproductive system: biparental transmission but haplodiploid-like inheritance in the mite Neoseiulus californicus. Heredity 84, 702–709 (2000). https://doi.org/10.1046/j.1365-2540.2000.00708.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2000.00708.x