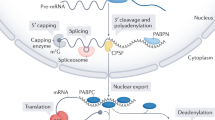
Abstract
A MAJOR pathway of messenger RNA degradation in eukaryotic cells is initiated by shortening of the poly(A) tail, which, at least in yeast, triggers a decapping reaction, thereby exposing the mRNA to 5′→ 3′ degradation1–4. Decapping is the key step in this decay pathway because the transcript body is rapidly degraded following decapping. Accordingly, decapping is the site of numerous controls, including inhibition of decapping by the poly (A) tail3,4 and modulation of mRNA decapping rate by specific sequences3–5. Moreover, a specialized decay pathway that degrades aberrant transcripts triggers rapid mRNA decapping independently of poly (A)-tail shortening6. We have identified a yeast gene, termed DCP1, that encodes the decapping enzyme, or an essential component of a decapping complex. The protein Dcpl is required for the normal decay of many unstable and stable yeast mRNAs, as well as mRNAs that are decapped independently of deadenylation. These results indicate that mRNA-specific rates of decapping, and thus decay, will result from differences in the interaction of the DCP1 decapping enzyme with individual transcripts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Decker, C. J. & Parker, R. Genes Dev. 7, 1632–1643 (1993).
Hsu, C. L. & Stevens, A. Molec. cell. Biol. 13, 4826–4835 (1993).
Muhlad, D., Decker, C. J. & Parker, R. Genes Dev. 8, 855–866 (1994).
Muhlrad, D., Decker, C. J. & Parker, R. Molec. cell. Biol. 15, 2145–2156 (1995).
Muhlrad, D. & Parker, R. Genes Dev. 6, 2100–2111 (1992).
Muhlrad, D. & Parker, R. Nature 370, 578–581 (1994).
Hatfield, L., Beelman, C. A., Stevens, A. & Parker, R. Molec. cell. Biol. (in the press).
Stevens, A. Molec. cell. Biol. 8, 2005–2010 (1988).
Hochuli, E., Döbeli, H. & Schacher, A. J. Chromatogr. 411, 177–184 (1987).
Beelman, C. A. & Parker, R. Cell 81, 179–183 (1995).
Decker, C. J. & Parker, R. Trends biochem. Sci. 19, 336–340 (1994).
Peltz, S. W., He, F., Welch, E. Jacobson, A. Prog. Nucleic Acids Res. molec. Biol. 17, 271–298 (1994).
He, F. & Jacobson, A. Genes Dev. 9, 437–454 (1995).
Caponigro, G. & Parker, R. Genes Dev. 9, 2421–2432 (1995).
Leeds, P., Peltz, S. W., Jacobson, A. & Culbertson, M. R. Genes Dev. 5, 2303–2314 (1991).
Leeds, P., Wood, J. M., Lee, B. -S. & Culbertson, M. R. Molec. cell. Biol. 12, 2165–2177 (1992).
Cui, Y., Hagan, K. W., Zhang, S. & Peltz, S. W. Genes Dev. 9, 423–436 (1995).
Johnson, A. W. & Kolodner, R. D. J. biol. Chem. 266, 14046–14054 (1991).
Mandart, E. & Parker, R. Molec. cell. Biol. 15, 6979–6986 (1995).
Harris, M. E. et al. EMBO J. 13, 3953–3963 (1994).
Caponigro, G., Muhlrad, D. & Parker, R. Molec. cell. Biol. 13, 5141–5148 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Beelman, C., Stevens, A., Caponigro, G. et al. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382, 642–646 (1996). https://doi.org/10.1038/382642a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/382642a0
This article is cited by
-
Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation
Nature Chemical Biology (2021)
-
Decapping enzyme 1A breaks X-chromosome symmetry by controlling Tsix elongation and RNA turnover
Nature Cell Biology (2020)
-
Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation
Cell Research (2014)
-
Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation
The EMBO Journal (2012)
-
Lsm1 promotes genomic stability by controlling histone mRNA decay
The EMBO Journal (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.