Abstract
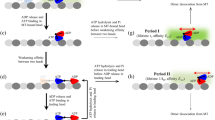
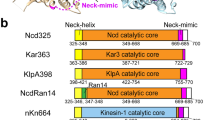
Kinesin motors are specialized enzymes that use hydrolysis of ATP to generate force and movement along their cellular tracks, the microtubules. Although numerous biochemical and biophysical studies have accumulated much data that link microtubule-assisted ATP hydrolysis to kinesin motion, the structural view of kinesin movement remains unclear. This study of the monomeric kinesin motor KIF1A combines X-ray crystallography and cryo-electron microscopy, and allows analysis of force-generating conformational changes at atomic resolution. The motor is revealed in its two functionally critical states—complexed with ADP and with a non-hydrolysable analogue of ATP. The conformational change observed between the ADP-bound and the ATP-like structures of the KIF1A catalytic core is modular, extends to all kinesins and is similar to the conformational change used by myosin motors and G proteins. Docking of the ADP-bound and ATP-like crystallographic models of KIF1A into the corresponding cryo-electron microscopy maps suggests a rationale for the plus-end directional bias associated with the kinesin catalytic core.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Okada, Y., Yamazaki, H., Sekine-Aizawa, Y. & Hirokawa, N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81, 769–780 (1995).
Hirokawa, N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279, 519–526 (1998).
Okada, Y. & Hirokawa, N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science 283, 152–157 (1999).
Kull, F. J., Sablin, E. P., Lau, R., Fletterick, R. J. & Vale, R. D. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380, 550–555 (1996).
Sablin, E. P., Kull, J. F., Cooke, R., Vale, R. D. & Fletterick, R. J. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature 380, 555–559 (1996).
Sack, S. et al. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry 36, 16155–16165 (1997).
Kozielski, F. et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91, 985–994 (1997).
Gulick, A., Song, H., Endow, S. & Rayment, I. X-ray crystal structure of the yeast KAR3 motor domain comlexed with MgADP to 2.3 Å resolution. Biochemistry 37, 1769–1776 (1998).
Sablin, E. P. et al. Direction determination in the minus-end-directed kinesin motor ncd. Nature 395, 813–816 (1998).
Kozielski, F., De Bonis, S., Burmeister, W. P., Cohen-Addad, C. & Wade, R. H. The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations. Struc. Fold Des. 7, 1407–1416 (1999).
Woehlke, G. et al. Microtubule interaction site of the kinesin motor. Cell 90, 207–216 (1997).
Sosa, H. et al. A model for the microtubule-ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell 90, 217–224 (1997).
Alonso, M., Damme, J., Vandekerckhove, J. & Cross, R. A. Proteolytic mapping of kinesin/ncd-microtubule interface: nucleotide-dependent conformational changes in the loops L8 and L12. EMBO J. 17, 945–951 (1998).
Hoenger, A. et al. Image reconstractions of microtubules decorated with monomeric and dimeric kinesins: comparison with X-ray structure and implications for motility. J. Cell Biol. 141, 419–430 (1998).
Kikkawa, M., Okada, Y. & Hirokawa, N. 15 Å resolution model of the monomeric kinesin motor, KIF1A. Cell 100, 241–252 (2000).
Okada, Y. & Hirokawa, N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl Acad. Sci. USA 97, 640–645 (2000).
Sprang, S. R. G protein mechanism: insights from structural analysis. Annu. Rev. Biochem. 66, 639–678 (1997).
Vale, R. D. Switches, latches, and amplifiers: common themes of molecular motors and G proteins. J. Cell Biol. 135, 291–302 (1996).
Rayment, I. et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261, 50–58 (1993).
Dominguez, R., Freyzon, Y., Trybus, K. M. & Cohen, C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell 94, 559–571 (1998).
Houdusse, A., Kalabokis, V. N., Himmel, D., Szent-Gyorgyi, A. G. & Cohen, C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell 97, 459–470 (1999).
Geeves, M. A. & Holmes, K. C. Structural mechanism of muscle contraction. Ann. Rev. Biochem. 68, 687–728 (1999).
Milburn, M. V. et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945 (1990).
Sack, S., Kull, F. J. & Mandelkow, E. Motor proteins of the kinesin family: structures, variations, and the nucleotide binding sites. Eur. J. Biochem. 262, 1–11 (1999).
Scheffzek, K. et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997).
Boriack-Sjodin, P. A., Margarit, S. M., Bar-Sagi, D. & Kuriyan, J. The structural basis of the activation of Ras by Sos. Nature 394, 337–343 (1998).
Kjeldgaard, M., Nyborg, J. & Clark, B. F. The GTP binding motif: variations on a theme. FASEB J. 10, 1347–1368 (1996).
Vale, R. D. & Milligan, R. A. The way things move: looking under the hood of molecular motor proteins. Science 288, 88–95 (2000).
Kjeldgaard, M., Nissen, P., Thirup, S. & Nyborg, J. The crystal structure of elongation factor Ef_Tu from Thermus aquaticus in the GTP conformation. Structure 1, 35–50 (1993).
Polekhina, G., Thirup, S., Kjeldgaard, M., Nissen, P. & Lippmann, C. Helix unwinding in the effector region of elongation factor Ef-Tu-GDP. Structure 4, 1141–1151 (1996).
Henningsen, U. & Schliwa, M. Reversal in the direction of movement of a molecular motor. Nature 389, 93–95 (1997).
Case, R. B., Pierce, D. W., Hom-Booher, N., Hart, C. & Vale, R. D. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 90, 959–966 (1997).
Endow, S. A. & Waligora, K. W. Determinants of kinesin motor polarity. Science 281, 1200–1202 (1998).
Vale, R. D., Case, R., Sablin, E., Hart, C. & Fletterick, R. Searching for kinesin's mechanical amplifier. Phil. Trans R. Soc. Lond. B 355, 449–457 (2000).
Rice, S. et al. A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 (1999).
Case, R. B., Rice, S., Hart, C. L., Ly, B. & Vale, R. D. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 10, 157–160 (2000).
Bohm, A., Gaudet, R. & Sigler, P. Structural aspects of heterotrimeric G-protein signalling. Curr. Opin. Biotech. 8, 480–487 (1997).
Hirose, K., Lockhart, A., Cross, R. A. & Amos, L. A. Nucleotide-dependent angular change in kinesin motor domain bound to tubulin. Nature 376, 277–279 (1995).
Hirose, K., Cross, R. A. & Amos, L. A. Nucleotide-dependent structural changes in dimeric ncd molecules complexed to microtubules. J. Mol. Biol. 278, 389–400 (1998).
Arnal, I. & Wade, R. H. Nucleotide-dependent conformations of the kinesin dimer interacting with microtubules. Structure 6, 33–38 (1998).
Hirose, K. et al. Structural comparison of dimeric Eg5, Neurospora kinesin (Nkin) and NCD head-Nkin neck chimera with conventional kinesin. EMBO J. 19, 5308–5314 (2000).
Nogales, E., Wolf, S. G. & Downing, K. H. Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 (1998).
Nogales, E., Whittaker, M., Milligan, R. A. & Downing, K. H. High-resolution model of the microtubule. Cell 96, 79–88 (1999).
Thorn, K. S., Ubersax, J. A. & Vale, R. D. Engineering the processive run length of the kinesin motor. J. Cell Biol. 151, 1093–1100 (2000).
Ma, Y.-Z. & Taylor, E. W. Interacting head mechanism of microtubule-kinesin ATPase. J. Biol. Chem. 272, 724–730 (1997).
Gilbert, S. P., Moyer, M. L. & Johnson, K. A. Alternating site mechanism of the kinesin ATPase. Biochemistry 37, 792–799 (1998).
Kawashima, T., Berthet-Colominas, C., Wulff, M., Cusak, S. & Leberman, R. The structure of the Escherichia coli EF-Tu-EF-Ts complex at 2.5 Å resolution. Nature 379, 511–518 (1996).
Song, H. & Endow, S. A. Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature 396, 587–590 (1998).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Brunger, A. T. et al. Crystallography and NMR system: a new software suit for macromolecular structure determination. Acta Cryst. D 54, 905–921 (1998).
Acknowledgements
We thank C. Sindelar and J. Turner for providing comments on this manuscript and sharing their results before publication. We also thank M. Sugaya for technical assistance. This work was supported by a Center of Excellence Grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (N.H.), and by funding from an NIH program project grant.
Author information
Authors and Affiliations
Corresponding author
Supplementary information

Figure 1 (GIF 8.9 KB)
Effective resolution of the cryo-EM maps. Fourier shell correlations between the two independent data sets are plotted. For the final map, the correlation coefficient fell to 0.5 at a resolution of 22 Å and 15 Å for the C351-ADP-microtubule complex and the C351-AMPPNP-microtubule complex, respectively.

Figure 2 (GIF 54.6 KB)
View showing electron density in the vicinity of the bound AMPPCP from a composite omit map (20 – 2.0 Å) contoured at 1.5 σ. A fragment of the final refined ATP-like KIF1A structure is shown. The γ-phosphate and Mg2+ ion (yellow asterisk) are labeled. The red asterisks indicate water molecules.
Rights and permissions
About this article
Cite this article
Kikkawa, M., Sablin, E., Okada, Y. et al. Switch-based mechanism of kinesin motors. Nature 411, 439–445 (2001). https://doi.org/10.1038/35078000
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35078000
This article is cited by
-
The role of kinesin family members in hepatobiliary carcinomas: from bench to bedside
Biomarker Research (2024)
-
A two-kinesin mechanism controls neurogenesis in the developing brain
Communications Biology (2023)
-
Kinesin-3 motors are fine-tuned at the molecular level to endow distinct mechanical outputs
BMC Biology (2022)
-
Genome wide analysis of kinesin gene family in Citrullus lanatus reveals an essential role in early fruit development
BMC Plant Biology (2021)
-
The macronuclear genome of the Antarctic psychrophilic marine ciliate Euplotes focardii reveals new insights on molecular cold adaptation
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.