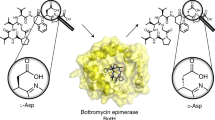
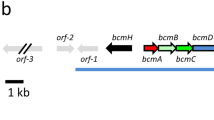
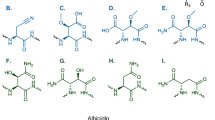
Abstract
ERYTHROMYCIN A, a clinically important polyketide antibiotic, is produced by the Gram-positive bacterium Saccharopolyspora erythraea.. In an arrangement that seems to be generally true of antibiotic biosynthetic genes in Streptomyces and related bacteria like S. erythraea1, the ery genes encoding the biosynthetic pathway to erythromvein are clustered around the gene (ermE) that confers self-resistance on S. erythraea2–6. The aglycone core of erythro-mycin A is derived from one propionyl-CoA and six methylmalonyl-CoA units, which are incorporated head-to-tail7–10 into the growing polyketide chain, in a process similar to that of fatty-acid biosynthesis1, to generate a macrolide intermediate, 6-deoxyeryth-ronolide B10. 6-Deoxyerythronolide B is converted into erythro-mycin A through the action2–5,10 of specific hydroxylases, glycosyItransferases and a methyltransferase. We report here the analysis of about 10 kilobases of DNA from S. erythraea, cloned by chromosome 'walking' outwards from the erythromycin-resistance determinant ermE, and previously shown to be essential for erythromycin biosynthesis5,11. Partial sequencing of this region12 indicates that it encodes the synthase. Our results confirm this, and reveal a novel organization of the erythromycin-producing polyketide synthase, which provides further insight into the mechanism of chain assembly.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Sherman, D. H. & Hopwood, D. A. A. Rev. Genet. 24, (in the press).
Stanzak, R., Matsushima, P., Baltz, R. H. & Rao, R. N. B. Bio/technology 4, 229–232 (1986).
Weber, J. M. & Losick, R. Gene 68, 173–180 (1988).
Weber, J. M., Wierman, C. K. & Hutchinson, C. R. J. Bact. 164, 425–433 (1985).
Weber, J. M., Leung, J. O., Maine, G. T., Potenz, R. H. B., Paulus, T. J. & DeWitt, J. P. J. Bact. 172, 2372–2383 (1990).
Dhillon, N., Hale, R. S., Cortes, J. & Leadlay, P. F. Molec. Microbiol. 3, 1405–1414 (1989).
Kaneda, T., Butte, J. C., Taubman, S. B. & Corcoran, J. W. J. biol. Chem. 237, 322–328 (1962).
Cane, D. E., Hasler, H. & Liang, T.-C. J. Am. chem. Soc. 103, 5960–5962 (1981).
Cane, D. E., Liang, T.-C., Taylor, P. B., Chang, C. & Yang, C.-C. J. Am. chem. Soc. 108, 4957–4964 (1986).
Seno, E. T. & Hutchinson, C. R. in The Bacteria Vol IX (eds Queener, S. W. & Day, L. E) 231–279 (Academic, New York, 1986).
Tuan, J. S. et al. Gene 90, 21–29 (1990).
Donadio, S. et al. In: Genetics and Microbiology of Industrial Microorganisms (eds Hershberger, C. L., Queener, S. W. & Hageman, G.) 53–59 (Am. Soc. of Microbiol., Washington, DC 1989).
Kauppinen, S., Siggaard-Andersen, M. & von Wettstein-Knowles, P. Carlsberg Res. Commun. 53, 357–370 (1988).
Bibb, M. J., Biro, S., Motamedi, H., Collins, J. F. & Hutchinson, C. R. EMBO J. 8, 2727–2736 (1989).
Sherman, D. H. et al. EMBO J. 8, 2727–2735 (1989).
Schweizer, M. et al. Molec. gen. genet. 203, 479–486 (1986).
Schweizer, M., Takabayashi, K., Laux, T., Beck, K. F. & Schreglmann, R. Nucleic Acids Res. 17, 567–586 (1988).
Chirala, S. S. et al. J. biol. Chem. 264, 3750–3757 (1989).
Mohamed, A. H., Chirala, S. S., Mody, N. H., Huang, W.-Y. & Wakil, S. J. J. biol. Chem. 263, 12315–12325 (1988).
Staden, R. Nucleic Acids Res. 12, 521–528 (1984).
Wakil, S. J. Biochemistry 28, 4523–4530 (1990).
Lawen, A. & Zocher, R. J. biol. Chem. 265, 11355–11360 (1990).
Skarpeid, H.-J., Zimmer, T.-L. & Von Doehren, H. Eur. J. Biochem. 189, 517–522 (1990).
Sanger, F., Nicklen, S. & Coulson, A. R. Proc. natn. Acad. Sci. U.S.A. 74, 5463–5467 (1977).
Frischauf, A.-M., Garoff, H. & Lehrach, H. Nucleic Acids Res. 8, 5541–5549 (1980).
Devereux, J., Haeberli, P. & Smithies, O. Nucleic Acids Res. 12, 387–395 (1984).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Cortes, J., Haydock, S., Roberts, G. et al. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature 348, 176–178 (1990). https://doi.org/10.1038/348176a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/348176a0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.