Abstract
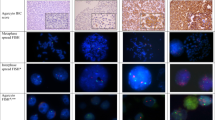
Chromosome 17q is frequently rearranged in breast cancer. Allelotyping studies have proposed the existence of at least four regions of allelic imbalance (AI). Here we present a study combining allelotyping using 19 CA repeat markers mapping in the 17q21 – 25 region and molecular cytogenetics (CGH and FISH). Allelotyping was undertaken on 178 pairs of cognate tumor and normal DNA in order to determine the number of regions of AI and define the shortest overlaps. AI ranged from 34 – 54% of the informative cases according to the marker and, overall, 66% of the tumors presented AI at one of the markers tested. Analysis of the patterns of imbalances revealed at least five common regions of imbalance respectively defined by markers: D17S855, which is intragenic of BRCA1 (SRO 1), D17S1607 (SRO 2), D17S1855 (SRO 3), between D17S789 and D17S785 (SRO 4) and D17S784 (SRO 5). In order to characterize the nature of the genetic events revealed by allelotyping we performed CGH analysis on a subset of 43 tumors presenting variable patterns of imbalance. CGH showed that AI at 17q could represent four different types of genetic events: loss of chromosome 17, gain of 17q, gain of 17q22 – q24, loss of 17q11 – q21 and/or 17q25 – qter. Some of these anomalies could occur concomitantly within the same tumor. Since 35% of the tumors analysed by CGH presented gains, these data indicated that AI at 17q were not solely indicative of losses of genetic material and could also represent DNA amplification. Gains were most commonly observed in the 17q23 – q24 regions. This suggested that AI in SRO 2 and SRO 3 corresponded to DNA amplification. To assess this, we isolated BAC clones by PCR screening for markers D17S1607 and D17S1855 and used these in FISH experiments on six breast tumor cell lines and 14 breast cancer specimens. FISH results showed that both D17S1607 and D17S1855 were frequently involved in DNA amplification (8 – 30 copies). Altogether, our data show that allelotyping can be efficiently used in amplicon mapping. Clinico-pathological correlations indicated that imbalance at 17q preferentially occurred in high grade, PR- and ERBB2 amplified tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bärlund M, Tirkkonen M, Forozan F, Tanner MM, Kallioniemi O and Kallioniemi A. . 1997 Genes Chromo. Cancer 20: 372–376.
Bautista S and Theillet C. . 1998 Genes Chromo. Cancer 22: 268–277.
Benitez J, Osorio A, Barroso A, Arranz E, Diaz-Guillen MA, Robledo M, Rodriguez de Cordoba S and Heine-Suner D. . 1997 Cancer Res. 57: 4217–4220.
Bugert P, Von Knobloch R and Kovacs G. . 1998 Int. J. Cancer 76: 337–340.
Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C and Theillet C. . 1997 Cancer Res. 57: 4360–4367.
Courjal F and Theillet C. . 1997 Cancer Res. 57: 4368–4377.
Cropp CS, Champeme MH, Lidereau R and Callahan R. . 1993 Cancer Res. 53: 5617–5619.
Devilee P, van Vliet M, Bardoel A, Kievits T, Kuipers Dijkshoorn N, Pearson PL and Cornelisse CJ. . 1991 Cancer Res. 51: 1020–1025.
Fletcher J. . (1994). In: Methods in Molecular Biology, Vol. 29: Chromosome Analysis Protocols. Godsen J.R. (ed.). Humana Press Inc: Totowa NJ pp 51–57.
Forozan F, Karhu R, Kononen J, Kallioniemi A and Kallioniemi OP. . 1997 Trends. Genet. 13: 405–409.
Foulkes WD, Black DM, Stamp GW, Solomon E and Trowsdale J. . 1993 Int. J. Cancer 54: 220–225.
Isola JJ, Kallioniemi OP, Chu LW, Fuqua SA, Hilsenbeck SG, Osborne CK and Waldman FM. . 1995 Am. J. Pathol. 147: 905–911.
Jacobs IJ, Smith SA, Wiseman RW, Futreal PA, Harrington T, Osborne RJ, Leech V, Molyneux A, Berchuck A, Ponder BAJ and Bast Jr RC. . 1993 Cancer Res. 53: 1218–1221.
Kalikin LM, Frank TS, Svoboda-Newman SM, Wetzel JC, Cooney KA and Petty EM. . 1997 Oncogene 14: 1991–1994.
Kenck C, Bugert P, Wilhelm M and Kovacs G. . 1997 Oncogene 14: 1093–1098.
Kuukasjarvi T, Tanner M, Pennanen S, Karhu R, Visakorpi T and Isola J. . 1997 Genes Chromo. Cancer 18: 94–101.
Mitelman F, Mertens F and Johansson B. . 1997 Nat. Genet. 15: 417–474.
Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B and Dutrillaux B. . 1994 Genes Chromo. Cancer 10: 160–170.
Munn KE, Walker RA, Menasce L and Varley JM. . 1996 Br. J. Cancer 73: 636–639.
Munn KE, Walker RA and Varley JM. . 1995 Oncogene 10: 1653–1657.
Niederacher D, Picard F, Van Roeyen C, An H-X, Bender HG and Beckmann MW. . 1997 Genes Chromo. Cancer 18: 181–192.
Plummer SJ, Adams L, Simmons JA and Casey G. . 1997a Oncogene 14: 2339–2345.
Plummer SJ, Paris MJ, Myles J, Tubbs R, Crowe J and Casey G. . 1997b Genes Chromo. Cancer 20: 354–362.
Plummer SJ, Simmons JA, Adams L and Casey G. . 1997c Genomics 45: 140–146.
Rajah R, Valentinis B and Cohen P. . 1997 J. Biol. Chem. 272: 12181–12188.
Ried T, Just KE, Holtgreve-Grez H, Du Manoir S, Speicher MR, Schröck E, Latham C, Blegen H, Zetterberg A, Cremer T and Auer G. . 1995 Cancer Res. 55: 5415–5423.
Theile M, Hartmann S, Scherthan H, Arnold W, Deppert W, Frege R, Glaab F, Haensch W and Scherneck S. . 1995 Oncogene 10: 439–447.
Wolf M, Aaltonen LA, Szymanska J, Tarkkanen M, Elomaa I and Knuutila S. . 1997 Cancer Genet. Cytogenet. 93: 33–38.
Acknowledgements
The authors are grateful to Dr Anne and Olli Kallioniemi for sharing unpublished observations with us, Dr Silvia Bautista for her help and advice for the molecular cytogenetics part of this work, Dr Laurent Journot for providing us the BAC pools for PCR screening. We wish to thank Professors Philippe Jeanteur, Henri Pujol and Jean-Bernard Dubois for their constant support. The expert technical support by Mrs Elisabeth Ursule and Annick Causse is gratefully acknowledged. This work was supported by funds from ARC, Ligue Nationale Contre le Cancer, FEGEFLUC. B Orsetti is supported by a doctoral fellowship from the comité de la Ligue de l'Hérault.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Orsetti, B., Courjal, F., Cuny, M. et al. 17q21-q25 aberrations in breast cancer: combined allelotyping and CGH analysis reveals 5 regions of allelic imbalance among which two correspond to DNA amplification. Oncogene 18, 6262–6270 (1999). https://doi.org/10.1038/sj.onc.1203006
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1203006
Keywords
This article is cited by
-
Genetic variability in the regulation of the expression cluster of MDR genes in patients with breast cancer
Cancer Chemotherapy and Pharmacology (2017)
-
Speckle-type POZ protein is negatively associated with malignancies and inhibits cell proliferation and migration in liver cancer
Tumor Biology (2015)
-
Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1
Oncogene (2011)
-
Na+/H+exchanger regulatory factor 1 inhibits platelet-derived growth factor signaling in breast cancer cells
Breast Cancer Research (2008)
-
Overexpression of the wip1 gene abrogates the p38 MAPK/p53/Wip1 pathway and silences p16 expression in human breast cancers
Breast Cancer Research and Treatment (2007)