Abstract
Published data on the associations between three well-characterized polymorphisms in the interleukin 6 and 10 (IL-6 and IL-10) genes and the risk of pneumonia are inconclusive. A meta-analysis was performed to derive a more precise estimate. The electronic databases MEDLINE (Ovid) and PubMed were searched from the earliest possible year to May 2014. A total of 9 articles met the criteria and these included 3460 patients with pneumonia and 3037 controls. The data were analyzed with RevMan software and risk estimates are expressed as odds ratios (ORs) and 95% confidence intervals (95% CIs). Analyses of the full data set failed to identify any significant association of pneumonia risk with the IL-6 gene -174C allele (OR = 1.00; 95% CI: 0.98–1.03), the IL-10 gene -592C allele (OR = 1.20; 95% CI: 0.95–1.52), or the IL-10 gene -1082A allele (OR = 1.21; 95% CI: 0.99–1.49). In a subgroup analysis by pneumonia type, ethnicity, sample size and quality score, no significantly increased risk of pneumonia was found for individuals carrying the IL-6 gene -174C allele. There was a low probability of publication bias, as reflected by the fail-safe number. This meta-analysis suggests that there is no significantly increased risk of pneumonia associated with previously reported IL-6 and IL-10 polymorphisms.
Similar content being viewed by others
Introduction
Pneumonia is a major cause of morbidity and mortality worldwide1. The strength of the immune response in humans is associated with the occurrence and severity of this disease. Cytokines released by inflammatory cells are important for the host immune response. Major pro-inflammatory cytokines include tumor necrosis factor-α (TNF-α) and interleukin 6 (IL-6). Interleukin 10 (IL-10) is considered to be the most important anti-inflammatory cytokine. Interleukin genes may play a key role in the pathogenesis of pneumonia2. The relationship between pneumonia and polymorphisms of interleukin genes has been studied extensively.
The importance of IL-6 in many physiological and pathological processes, particularly in the inflammatory response, has been reported3. In patients with unilateral pneumonia, Dehoux and colleagues found that the IL-6 level in bronchoalveolar lavage fluid obtained from the infected lung was significantly higher than that in the uninfected side or in the plasma. Waage et al. showed that elevated plasma levels of IL-6 are associated with high mortality4. In addition, several studies have reported increased IL-10 levels in the blood of patients with severe sepsis or septic shock5.
Several polymorphisms in the promoter regions of IL-6 and IL-10, such as IL-6 -174G/C (rs1800795), IL-6 -572G/C (rs1800796), IL-10 -592C/A (rs1800872) and IL-10 -1082G/A (rs1800896), have been identified. Previous studies have reported associations between IL-6 and IL-10 polymorphisms and the risk of pneumonia6,7,8,9,10,11,12,13,14. Although exhaustive association studies have been undertaken to address this issue, no definitive conclusion has yet been reached and the results have been irreproducible. To generate more information, we carried out a meta-analysis of all of the available case-control studies to investigate the association of genetic polymorphisms of IL-6 and IL-10 with the risk of pneumonia. The selection of polymorphisms under investigation was straightforward if three or more unduplicated studies were available for a certain polymorphism of IL-6 and IL-10 genes.
Methods
Ethics
The study protocol was approved by the Coordinating Ethics Committee of Ruijin Hospital and the study methods were carried out in accordance with the approved guidelines.
Search strategy for the identification of studies
We searched PubMed and MEDLINE (Ovid) for articles published before May 2014. The subject terms included either interleukin-6 (or IL-6) or interleukin-10 (or IL-10) and pneumonia. The search results were expressed using Boolean operators: ((interleukin-6) OR IL-6 OR (interleukin-10) OR IL-10) AND (pneumonia) AND (gene OR polymorphism OR alleles OR variants)) AND English [Language].
Inclusion/exclusion criteria
Our analyses were restricted to articles that fulfilled the following inclusion criteria (with all having to be satisfied): 1) investigation of the association between genetic polymorphisms of the IL-6 and IL-10 genes and pneumonia among unrelated subjects; 2) genotypes of the examined polymorphisms were tested in a validated sample size; 3) a case-control study design; and 4) sufficient information on the genotypes or alleles of the examined polymorphisms to allow estimation of the odds ratio (OR) and its corresponding 95% confidence interval (95% CI). Articles were excluded (with one condition being sufficient to do so) if they investigated the progression or severity of pneumonia, phenotype modification, or response to treatment or survival, as well as if they were conference abstracts, case reports/series, editorials, review articles, or non-English articles. If there were multiple publications from the same study group, the most complete and recent results were used. The search results were limited to articles published in English and studies performed in humans.
Data extraction
Two reviewers (C.H. and L.N.) independently assessed all potentially relevant studies and reached a consensus on all items. In cases of disagreement, a third author provided an assessment. The following data were collected from each study: first author, year of publication, ethnicity, study design, diagnostic criteria, baseline characteristics of the study population, total number of cases and controls and genotype distributions in cases and controls. After data extraction, discrepancies were adjudicated by discussion until a consensus was reached.
Quality score assessment
The study quality was evaluated using a quality assessment score developed for genetic association studies by Thakkinstian and colleagues15. Total scores ranged from 0 (the worst) to 12 (the best). The criteria for the quality assessment of genetic associations between the IL-6 gene C-174G polymorphism and pneumonia are described in Table S1.
Statistical methods
The meta-analysis was calculated using Review Manager version 5.0.19 software, available at http://ims.cochrane.org/revman/download. The Hardy-Weinberg equilibrium was assessed using Pearson's χ2 test or Fisher’s exact test (SAS version 9.1.3, Institute Inc., Cary, NC, USA). The inconsistency index (I2) was used to quantify the presence of between-study heterogeneity, with statistical significance set at 0.116. When the P value was >0.10, the pooled OR was calculated using the fixed-effects model; otherwise, a random-effects model was used. Sensitivity analyses were performed to look at more narrowly drawn subsets of the studies by removing an individual study or by removing studies with similar feature to assess their influence separately. Predefined subgroup analyses were performed a priori according to ethnicity (Caucasian or mixed), age (adult or pediatric), the pneumonia type (CAP or HAP), total sample size (<500 subjects or ≥500 subjects), or the quality score (score <11 or score ≥11).
Publication bias was assessed by the fail-safe number (Nfs), with the significance set at 0.05 for each meta-comparison. Specifically, if the calculated Nfs value was smaller than the number of studies observed, the meta-analysis results might have publication bias. We calculated the Nfs0.05 according to the formula Nfs0.05 = (ΣZ/1.64) 2 − k, where k is the number of articles included.
Results
Study characteristics
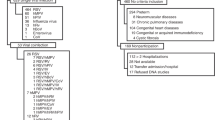
Based on the search strategy, our primary search produced 39 potentially relevant articles, of which 9 articles met the inclusion criteria6,7,8,9,10,11,12,13,14. In total, 3460 patients with pneumonia and 3037 controls were examined. The detailed selection process is presented in Figure 1. The details of each excluded study have been uploaded as a supplementary information file (Table S2). The baseline characteristics of the included studies are presented in Table 1.
Of these studies, 7 articles examined the association of the IL-6 -174G/C polymorphism with pneumonia6,7,8,9,10,11,12. Because Salnikova LE et al. had published two articles on the same study group, we used the more recent result10,17. Three articles focused on the IL-10 gene -592C/A polymorphism8,11,13 and 3 articles focused on the IL-10 gene -1082G/A polymorphism8,11,14.
Overall analyses
Figure 2 depicts the pooled risk estimates of developing pneumonia for the mutant alleles of the three IL-6 and IL-10 gene polymorphisms. Under a random-effects model, the analyses of the full data set failed to reveal any significant association of the IL-6 -174C allele (OR = 1.00; 95% CI: 0.93–1.08), the IL-10 -592C allele (OR = 1.20; 95% CI: 0.95–1.52), or the IL-10 -1082A allele (OR = 1.43; 95% CI: 0.76–2.70) with risk of pneumonia. Sensitivity analyses were performed by excluding studies with controls not in HWE. The results show that the associations between the IL-10 gene -592C and IL-10 gene -1082A polymorphisms and pneumonia risk were not significantly altered.
Subgroup analyses
In view of the number of included articles, subgroup analyses were undertaken only for the IL-6 gene -174 C/G polymorphism, with regard to age, pneumonia type, ethnicity, sample size and quality score (table 2). The subgroup analysis stratified by age showed that no associations existed in adults (OR = 1.02, 95% CI: 0.95–1.11, p = 0.56). In the subgroup analysis of the type of pneumonia, no significantly increased risk of pneumonia was found for CAP (OR = 1.00, 95% CI: 0.93–1.08, p = 0.93). The subgroup analysis stratified by ethnicity showed that no association existed in Caucasians (OR = 1.02, 95% CI: 0.95–1.11, p = 0.56). With regard to sample size, no significance was reached in large studies (the total sample size ≥500 participants) or in small studies (the total sample size <500 participants). With regard to quality score, there were no significant findings observed under any of the four genetic models in low-quality studies (quality score <11) or in high-quality studies (quality score ≥11).
Publication bias
The Nfs values were calculated to assess the potential existence of publication bias. At a significance level of 0.05, the Nfs0.05 values were consistently greater than the number of studies included in this meta-analysis for all polymorphisms under investigation. In the analysis of the IL-6 gene -174 C/G polymorphism and pneumonia risk, the resultant symmetrical funnel shape was consistent with the absence of publication bias in the funnel plot for contrasts of C versus G (P-Egger test = 0.475) (Figure 3).
Begg’s funnel plot of the Egger test for publication bias of the IL-6 gene -174 C/G polymorphism and pneumonia risk analysis.
The horizontal line in the funnel plot indicates the fixed-effects summary estimates and the sloping lines indicate the expected 95% CI for a given standard error. The sizes of the circles in each plot are positively proportional to the sample sizes of each study.
Discussion and conclusions
In this study, we sought to investigate the association of IL-6 and IL-10 genetic polymorphisms with pneumonia risk by conducting a meta-analysis of studies reported in English journals and we included 9 articles covering 6497 subjects. This meta-analysis demonstrated an absence of association between the IL-6 gene C-174G, IL-10 gene C-592A and IL-10 gene G-1082A polymorphisms and pneumonia risk. Moreover, a subgroup analysis indicated no significantly increased risk of CAP among adults. To the authors’ knowledge, this is the first meta-analysis investigating the association of the IL-6 gene C-174G, IL-10 gene C-592A and IL-10 gene G-1082A genetic polymorphisms with pneumonia risk.
Results from our meta-analysis show a lack of association between IL-6 and IL-10 gene polymorphisms and pneumonia risk. Although many studies have reported that the allele IL-6-174C is associated with increased IL-6 secretion18,19, our study did not find such an association. Some studies have shown that IL-10-1082 G is associated with increased secretion of IL-10 in chronic hepatitis B virus infection20 and clinical malaria21, although no significantly increased risk of pneumonia was found. There are two potential reasons for the results. First, because of the complex nature of pneumonia, it is unlikely that a single nucleotide polymorphism in a single gene would be associated with an increased risk of pneumonia or mortality, without a contribution from other polymorphic susceptibility genes. Second, other factors, such as age, pathogenic organism, medical treatment and nutrient status, can also influence the development or the prognosis of pneumonia. Three studies have reported on the association of IL-6 -174 GG genotype with systemic inflammatory response syndrome (SIRS) and mortality from pneumonia7,9,22. However, the studies used different standards to extract data, so a meta-analysis could not be conducted. This issue needs to be further studied. Paats et al. found that IL-6 and IL-10 play important roles in CAP. They showed that the level of IL-6 was significantly increased in the bronchoalveolar lavage fluid of CAP patients compared with healthy individuals and that serum levels of IL-6 and IL-10 were significantly higher in patients with severe CAP than in those with non-severe CAP or healthy individuals23. Kwan J and colleagues found that IL-6 is independently associated with stroke-associated infection and may be a key biomarker24.
We also carried out subgroup analyses by age, pneumonia type, ethnicity, sample size and quality score. For ethnicity, our results showed no significant increase in risk of pneumonia among Caucasians. Subgroup analyses also did not detect a significant association between IL-6 -174 and pneumonia risk in adults with CAP. We also found that no association existed between IL-6 -174 and pneumonia for any sample size or quality score.
This study has several limitations. First, only published studies in English were included; it is possible that some relevant published or unpublished studies with null results were missed, which might have biased the results. Second, owing to the relatively small number of eligible studies, we were unable to perform further subgroup analyses, such as those by ethnicity or gender, because of limited data. Third, because the data extracted from the primary publications were insufficient, we could not assess the effects of the IL-6 -572G/C, 1753C/G, 2954G/C, IL-10 -819 C/T and interleukin-1 receptor antagonist intron 225 polymorphisms on pneumonia risk. Fourth, the statistical heterogeneities of the effects of IL-10 C-592A and IL-10 G-1082A were significant in our meta-analyses, likely because only three studies of these polymorphisms were included in the meta-analysis and these studies were conducted in different countries and had different sample sizes. Finally, the lack of original data in the eligible studies limited the evaluation of the effects of gene-gene interactions in pneumonia. Therefore, the jury remains out before the eventual truth prevails. We minimized the likelihood of bias by creating a detailed protocol before initiating our study, performing a meticulous search for publications and using explicit methods for publication selection, data extraction and analysis.
In conclusion, our results suggest that IL-6 and IL-10 gene polymorphisms are not associated with the risk of pneumonia. Future studies with large sample sizes and more ethnic groups are needed to confirm our findings. Moreover, other interleukin polymorphisms and gene-gene interactions should also be considered in future studies.
References
Garau, J. et al. Factors impacting on length of stay and mortality of community-acquired pneumonia. Clin Microbiol Infect. 14, 322–329 (2008).
Wunderink, R. G. & Waterer, G. W. Genetics of community-acquired pneumonia. Semin Respir Crit Care Med. 26, 553–562 (2005).
Zobel, K. et al. Interleukin 6, lipopolysaccharide-binding protein and interleukin 10 in the prediction of risk and etiologic patterns in patients with community-acquired pneumonia: results from the German competence network CAPNETZ. BMC Pulm Med. 12, 6 (2012).
Waage, A., Brandtzaeg, P., Halstensen, A., Kierulf, P. & Espevik, T. The complex pattern of cytokines in serum from patients with meningococcal septic shock. Association between interleukin 6, interleukin 1 and fatal outcome. J Exp Med. 169, 333–338 (1989).
Friedman, G. et al. Blood interleukin 10 levels parallel the severity of septic shock. J Crit Care. 12, 183–187 (1997).
Schaaf, B. et al. The interleukin-6 -174 promoter polymorphism is associated with extrapulmonary bacterial dissemination in Streptococcus pneumoniae infection. Cytokine 31, 324–328 (2005).
Sole-Violan, J. et al. Genetic variability in the severity and outcome of community-acquired pneumonia. Respir Med. 104, 440–447 (2010).
Endeman, H. et al. Systemic cytokine response in patients with community-acquired pneumonia. Eur Respir J. 37, 1431–1438 (2011).
Martin-Loeches, I. et al. Variants at the promoter of the interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intensive Care Med. 38, 256–262 (2012).
Salnikova, L. E., Smelaya, T. V., Moroz, V. V., Golubev, A. M. & Rubanovich, A. V. Functional polymorphisms in the CYP1A1, ACE and IL-6 genes contribute to susceptibility to community-acquired and nosocomial pneumonia. Int J Infect Dis. 17, e433–442 (2013).
Martinez-Ocana, J. et al. Plasma cytokine levels and cytokine gene polymorphisms in Mexican patients during the influenza pandemic A(H1N1)pdm09. J Clin Virol. 58, 108–113 (2013).
Zidan, H. E., Elbehedy, R. M. & Azab, S. F. IL6-174 G/C gene polymorphism and its relation to serum IL6 in Egyptian children with community-acquired pneumonia. Cytokine 67, 60–64 (2014).
Wan, Q. Q., Li, J. L., Ye, Q. F. & Zhou, J. D. Genetic association of tumor necrosis factor-beta, interleukin-10 and interleukin-1 gene cluster polymorphism with susceptibility to pneumonia in kidney transplant recipients. Transplant Proc. 45, 2211–2214 (2013).
Schaaf, B. M. et al. Pneumococcal septic shock is associated with the interleukin-10-1082 gene promoter polymorphism. Am J Respir Crit Care Med. 168, 476–480 (2003).
Thakkinstian, A. et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol 162, 201–211 (2005).
Bowden, J., Tierney, J. F., Copas, A. J. & Burdett, S. Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol 7, 41(2011).
Salnikova, L. E., Smelaya, T. V., Moroz, V. V., Golubev, A. M. & Rubanovich, A. V. Host genetic risk factors for community-acquired pneumonia. Gene. 518, 449–456 (2013).
Muller-Steinhardt, M., Ebel, B. & Hartel, C. The impact of interleukin-6 promoter -597/-572/-174 genotype on interleukin-6 production after lipopolysaccharide stimulation. Clin Exp Immunol 147, 339–345 (2007).
Tischendorf, J. J. et al. The interleukin-6 (IL6)-174 G/C promoter genotype is associated with the presence of septic shock and the ex vivo secretion of IL6. Int J Immunogenet 34, 413–418 (2007).
Wu, J. F. et al. Serum levels of interleukin-10 and interleukin-12 predict early, spontaneous hepatitis B virus eantigen seroconversion. Gastroenterology 138, 165–172 (2010).
Zhang, G. et al. Interleukin-10 (IL-10) Polymorphisms Are Associated with IL-10 Production and Clinical Malaria in Young Children. Infect Immun 80, 2316–2322 (2012).
Gallagher, P. M. et al. Association of IL-10 polymorphism with severity of illness in community acquired pneumonia. Thorax 58, 154–156 (2003).
Paats, M. S. et al. Local and systemic cytokine profiles in nonsevere and severe community-acquired pneumonia. Eur Respir J. 41, 1378–1385 (2013).
Kwan, J. et al. IL-6 is a predictive biomarker for stroke associated infection and future mortality in the elderly after an ischemic stroke. Exp Gerontol 48, 960–965 (2013).
Patwari, P. P. et al. Interleukin-1 receptor antagonist intron 2 variable number of tandem repeats polymorphism and respiratory failure in children with community-acquired pneumonia. Pediatr Crit Care Med. 9, 553–559 (2008).
Author information
Authors and Affiliations
Contributions
Conception and design of the experiments: Y.F. and G.C.S. Execution of the experiments: H.C. and N.L. Analysis of the data: Y.F., H.Y.W. and Q.J.C. Contribution of reagents/materials/analytical tools: Y.F. and G.C.S. Composition of the manuscript: Y.F. and Q.J.C.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
This study was supported by the National Natural Science Foundation of China (81201837).
Electronic supplementary material
Supplementary Information
Table S1,S2
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, H., Li, N., Wan, H. et al. Associations of Three Well-Characterized Polymorphisms in the IL-6 and IL-10 Genes with Pneumonia: A Meta-Analysis. Sci Rep 5, 8559 (2015). https://doi.org/10.1038/srep08559
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08559
This article is cited by
-
Genetic dissection of host immune response in pneumonia development and progression
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.