Abstract
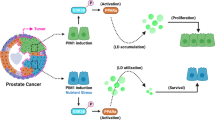
Lipid droplet tethering with mitochondria for fatty acid oxidation is critical for tumor cells to counteract energy stress. However, the underlying mechanism remains unclear. Here, we demonstrate that glucose deprivation induces phosphorylation of the glycolytic enzyme phosphofructokinase, liver type (PFKL), reducing its activity and favoring its interaction with perilipin 2 (PLIN2). On lipid droplets, PFKL acts as a protein kinase and phosphorylates PLIN2 to promote the binding of PLIN2 to carnitine palmitoyltransferase 1A (CPT1A). This results in the tethering of lipid droplets and mitochondria and the recruitment of adipose triglyceride lipase to the lipid droplet–mitochondria tethering regions to engage lipid mobilization. Interfering with this cascade inhibits tumor cell proliferation, promotes apoptosis and blunts liver tumor growth in male mice. These results reveal that energy stress confers a moonlight function to PFKL as a protein kinase to tether lipid droplets with mitochondria and highlight the crucial role of PFKL in the integrated regulation of glycolysis, lipid metabolism and mitochondrial oxidation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The human HCC data for PFKL expression were obtained from The Cancer Genome Atlas Research Network (https://ualcan.path.uab.edu/analysis.html). Homology models of PFKL were generated using the SWISS-MODEL web server (https://swissmodel.expasy.org) based on the crystal structure of human PFKP bound to ATP-Mg2+, retrieved from the Protein Data Bank (PDB 4XYJ). The dataset for the phosphorylated level of PLIN2 at Ser159 was sourced from the Cancer Proteogenomic Data Analysis Site (https://cprosite.ccr.cancer.gov). The wild-type structure of PLIN2 was predicted using AlphaFold (https://alphafold.ebi.ac.uk/entry/Q99541). For mass spectrometry analysis, proteins were identified by matching their fragment spectra against the Swiss-Prot database (http://www.expasy.ch/sprot). The data that support the findings of this study are available within the paper and Supplementary Information. Source data for Figs. 1–5 and Extended Data Figs. 1–7 are provided as Source Data files. Source data are provided with this paper.
References
Walther, T. C. & Farese, R. V. Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 (2012).
Olzmann, J. A. & Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019).
Sztalryd, C. & Brasaemle, D. L. The perilipin family of lipid droplet proteins: gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1221–1232 (2017).
Henne, W. M., Reese, M. L. & Goodman, J. M. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 37, e98947 (2018).
Benador, I. Y., Veliova, M., Liesa, M. & Shirihai, O. S. Mitochondria bound to lipid droplets: where mitochondrial dynamics regulate lipid storage and utilization. Cell Metab. 29, 827–835 (2019).
Kaushik, S. & Cuervo, A. M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015).
Wu, X. et al. Lipid droplets maintain energy homeostasis and glioblastoma growth via autophagic release of stored fatty acids. iScience 23, 101569 (2020).
Liu, R. et al. Choline kinase alpha 2 acts as a protein kinase to promote lipolysis of lipid droplets. Mol. Cell 81, 2722–2735.e9 (2021).
Zhu, R. et al. Choline kinase alpha2 promotes lipid droplet lipolysis in non-small-cell lung carcinoma. Front. Oncol. 12, 848483 (2022).
Ma, Q., Meng, Z., Meng, Y., Liu, R. & Lu, Z. A moonlighting function of choline kinase alpha 2 in the initiation of lipid droplet lipolysis in cancer cells. Cancer Commun. (Lond) 41, 933–936 (2021).
Rambold, A. S., Cohen, S. & Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015).
Benador, I. Y. et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869–885.e6 (2018).
Wang, H. et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52, 2159–2168 (2011).
Granneman, J. G., Moore, H. P., Mottillo, E. P., Zhu, Z. & Zhou, L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 286, 5126–5135 (2011).
Boutant, M. et al. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 36, 1543–1558 (2017).
Freyre, C. A. C., Rauher, P. C., Ejsing, C. S. & Klemm, R. W. MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell 76, 811–825.e14 (2019).
Fan, T. et al. Metabolomic and transcriptomic profiling of hepatocellular carcinomas in Hras12V transgenic mice. Cancer Med. 6, 2370–2384 (2017).
van der Leij, F. R., Huijkman, N. C., Boomsma, C., Kuipers, J. R. & Bartelds, B. Genomics of the human carnitine acyltransferase genes. Mol. Genet. Metab. 71, 139–153 (2000).
Lee, J. H. et al. EGFR-phosphorylated platelet isoform of phosphofructokinase 1 promotes PI3K activation. Mol. Cell 70, 197–210.e7 (2018).
Lee, J. H. et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat. Commun. 8, 949 (2017).
Olarte, M. J. et al. Determinants of endoplasmic reticulum-to-lipid droplet protein targeting. Dev. Cell 54, 471–487.e7 (2020).
Webb, B. A. et al. Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature 523, 111–114 (2015).
Lu, Z. & Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 43, 301–310 (2018).
Li, X., Egervari, G., Wang, Y., Berger, S. L. & Lu, Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol. 19, 563–578 (2018).
Xu, D. et al. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metab. 33, 33–50 (2021).
Guo, D. et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IϰBα. Cell Metab. 34, 1312–1324.e6 (2022).
Webb, B. A., Dosey, A. M., Wittmann, T., Kollman, J. M. & Barber, D. L. The glycolytic enzyme phosphofructokinase-1 assembles into filaments. J. Cell Biol. 216, 2305–2313 (2017).
Bell, M. et al. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes 57, 2037–2045 (2008).
Wang, Z. et al. Glycolysis inhibitor 2-deoxy-d-glucose suppresses carcinogen-induced rat hepatocarcinogenesis by restricting cancer cell metabolism. Mol. Med. Rep. 11, 1917–1924 (2015).
Pol, A., Gross, S. P. & Parton, R. G. Biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204, 635–646 (2014).
Bian, X. et al. Regulation of gene expression by glycolytic and gluconeogenic enzymes. Trends Cell Biol. 32, 786–799 (2022).
Lee, J. H. et al. Phosphofructokinase 1 platelet isoform promotes β-catenin transactivation for tumor development. Front. Oncol. 10, 211 (2020).
Long, M. & McWilliams, T. G. Lipid droplets promote efficient mitophagy. Autophagy 19, 724–725 (2023).
Nguyen, T. B. et al. DGAT1-Dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21.e5 (2017).
MacPherson, R. E., Ramos, S. V., Vandenboom, R., Roy, B. D. & Peters, S. J. Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 304, R644–R650 (2013).
Najt, C. P. et al. Organelle interactions compartmentalize hepatic fatty acid trafficking and metabolism. Cell Rep. 42, 112435 (2023).
Pollak, N. M. et al. The interplay of protein kinase A and perilipin 5 regulates cardiac lipolysis. J. Biol. Chem. 290, 1295–1306 (2015).
Sieradzki, J., Ksiezyk, M. & Korzeniowska, D. Insulin secretion in acromegaly and Cushing’s syndrome. Endokrynol. Pol. 38, 407–413 (1987).
Miner, G. E. et al. PLIN5 interacts with FATP4 at membrane contact sites to promote lipid droplet-to-mitochondria fatty acid transport. Dev. Cell 58, 1250–1265.e6 (2023).
Ouyang, Q. et al. Rab8a as a mitochondrial receptor for lipid droplets in skeletal muscle. Dev. Cell 58, 289–305.e6 (2023).
Wang, H. et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286, 15707–15715 (2011).
Yang, W. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 (2012).
Li, X. et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell 61, 705–719 (2016).
Li, X. et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat. Cell Biol. 18, 561–571 (2016).
Ding, Y. et al. Isolating lipid droplets from multiple species. Nat. Protoc. 8, 43–51 (2013).
Ji, H. T. et al. EGF-induced ERK activation promotes CK2-mediated disassociation of α-catenin from β-catenin and transactivation of β-catenin. Mol. Cell 36, 547–559 (2009).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Wang, Z. et al. Fructose-1,6-bisphosphatase 1 functions as a protein phosphatase to dephosphorylate histone H3 and suppresses PPARα-regulated gene transcription and tumour growth. Nat. Cell Biol. 24, 1655–1665 (2022).
Fernandes, P. M., Kinkead, J., McNae, I., Michels, P. A. M. & Walkinshaw, M. D. Biochemical and transcript level differences between the three human phosphofructokinases show optimisation of each isoform for specific metabolic niches. Biochem. J. 477, 4425–4441 (2020).
Park, H. et al. PAAN/MIF nuclease inhibition prevents neurodegeneration in Parkinson’s disease. Cell 185, 1943–1959.e21 (2022).
Jia, Y., Quinn, C. M., Kwak, S. & Talanian, R. V. Current in vitro kinase assay technologies: the quest for a universal format. Curr. Drug Discov. Technol. 5, 59–69 (2008).
Sanghera, J., Li, R. & Yan, J. Comparison of the luminescent ADP-Glo assay to a standard radiometric assay for measurement of protein kinase activity. Assay Drug Dev. Technol. 7, 615–622 (2009).
Wang, J. et al. An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D-TSG101 interactions. Nat. Commun. 12, 1252 (2021).
Yang, W. et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122 (2011).
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (82188102, 82030074, Z.L.; 82203553, Y.M.; 82372816, G.D.; 82072630, D.X.; 82372814, 82173114, Z.W.), the Ministry of Science and Technology of the People’s Republic of China (2020YFA0803300, Z.L.; 2021YFA0805600, D.X.), Zhejiang Natural Science Foundation Key Project (LD22H160002, D.X.), Zhejiang Natural Science Foundation Discovery Project (LQ22H160023, Z.W.), Taishan Scholar Project from Shandong province (tsqn202312174, P.Y.), and the China Postdoctoral Science Foundation (2021M700123, Y.M.). Z.L. is the Kuancheng Wang Distinguished Chair. We thank X. Hong from the Core Facilities of Zhejiang University School of Medicine for her technical support, L. Wang and P. Yang from the Center of Cryo-Electron Microscopy at Zhejiang University for their technical assistance with transmission electron microscopy and the Metabolomics Facility Center of Metabolomics and Lipidomics in the National Protein Science Technology Center of Tsinghua University for LC–MS/MS experiments.
Author information
Authors and Affiliations
Contributions
Z.L. conceptualized the study. Z.L., D.X. and Y.M. acquired the funding support and supervised the study. Y.M., D.G. and L.L. performed the experiments and statistical analysis; H.Z. performed the molecular dynamics simulation and peptide docking analysis. W.X., Shudi L., X.J., Shan L., X.H., R.Z., R.S., L.X., Q.W., H.H., J.T., H.J., Z.W, P.Y. and D.X. provided reagents, technical support and conceptual advice. Z.L. and Y.M. wrote the paper with comments from all authors.
Corresponding authors
Ethics declarations
Competing interests
Z.L. owns shares in Signalway Biotechnology, which supplied the rabbit antibodies that recognize PFKL pT331 and PLIN2 pS159. Z.L.’s interest in this company had no bearing on it being chosen to supply these reagents. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Bradley Webb, Deliang Guo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Alfredo Giménez-Cassina, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 PFKL is required for the interaction between PLIN2 and CPT1A, lipid droplet-mitochondria tethering, and lipid droplet lipolysis.
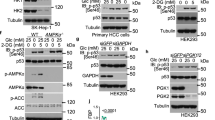
b, c, e, g, j, Immunoblotting analyses with the indicated antibodies were performed, presenting representative results from three independent experiments. d, i, k, l, Representative images from three independent experiments. a, LDs from Huh7 cells were purified after 40 min of glucose deprivation and subjected to LC-MS/MS. Selected peptide hits of LD-associated proteins are shown. b, LM3 cells were treated with 40 min of glucose deprivation. c, Huh7 cells with depleted endogenous PLIN2 and reconstituted expression of Flag-rPLIN2 protein were treated with glucose deprivation for the indicated period of time. d, Representative TEM images of Huh7 cells treated with 1 h of glucose deprivation were presented (left). Percentages of mitochondria-associated LDs per field were quantified. n = 15 or 16 microscope fields (right). e, Huh7 cells were infected with lentiviruses expressing shCtrl or shCPT1A. f, Huh7 cells with glucose deprivation for the indicated period of time were subjected to glycolytic metabolites analysis by LC-MS/MS. n = 3. g, Huh7 cells expressing Flag-PLIN2 were treated with 40 min of glucose deprivation. Cell lysates were incubated with glycolytic metabolites (F6P, FBP, DHAP, G3P, 3PG) followed by immunoprecipitation. h, PFKL expression levels were analyzed in normal liver and HCC tissues using a TCGA dataset. Normal: minimum, 22.235; maximum, 83.356; lower quartile, 38.085; upper quartile, 61.781; and median 48.352; HCC: minimum, 13.449; maximum, 166.926; lower quartile, 52.154; upper quartile, 98.358; and median, 72.891. The horizontal lines mark the median. n (Normal) = 50, n (HCC) = 371. i, Workflow for purifying LDs from Huh7 cells expressing GFP-Flag-LiveDrop (left). Immunofluorescence analyses were performed (right). j, LM3 cells stably expressing shCtrl or shPFKL were transfected with Flag-PLIN2 and were treated with 40 min of glucose deprivation (left). PFKL depletion efficiency was quantified. n = 3 (right). k, LM3 cells stably expressing shCtrl or shPFKL#1 were stimulated by 40 min of glucose deprivation (left). Percentages of mitochondria-associated LDs per cell were quantified. n (shCtrl) = 20, n (shPFKL) = 20 (right). l, m, LM3 cells stably expressing shCtrl or shPFKL#1 were treated with 4 h glucose deprivation for (l). LDs number and the percentage of LDs area per cell were quantified. n (shCtrl, Glc+) = 30, n (shCtrl, Glc‒) = 30, n (shPFKL, Glc‒) = 30 (m). d, f, h, j, k, m, Data are presented as means ± SD of three biologically independent replicates, analyzed by two-sided unpaired Student’s t-test (h, j, k) or one-way ANOVA (m).
Extended Data Fig. 2 p38-mediated PFKL T331 phosphorylation is necessary for PFKL binding to PLIN2.
a, b, e, g, h, i, Immunoblotting analyses with the indicated antibodies were performed, presenting representative results from three independent experiments. a, Huh7 cells were treated with or without the indicated inhibitors for 30 min before 40 min of glucose deprivation. b, Huh7 cells expressing Flag-p38 or a Flag vector were treated with 40 min of glucose deprivation. Immunoprecipitation analyses with an anti-Flag antibody were performed. c, An in vitro phosphorylation assay was performed by mixing bacterially purified His-SUMO-PFKL with active His-p38. Liquid chromatography-tandem mass spectrometry/mass spectrometry (LC-MS/MS) analyses showed a tryptic fragment at m/z 796.90723 Da (+13.52 mmu/+16.97 ppm), which was matched with the +4 charged peptide 316- MGMEAVMALLEATPD(pT)PACVVTLSGNQSVR-345 of PFKL, suggesting that PFKL T331 was phosphorylated. The Xcorr score was 3.07. d, Alignment of protein sequences spanning PFKL T331 in different species. Human PFKL T331 and the corresponding sites in other species are shown in red. e, Huh7 cells stably expressing shCtrl or shPFKL#1 were treated with 40 min of glucose deprivation. f, Immunohistochemical analyses of human HCC samples were performed with the indicated antibodies in the presence or absence of a blocking peptide for PFKL pT331. Representative images from three independent experiments are shown. g, WT Flag-rPFKL or Flag-rPFKL T331A was expressed in Huh7 cells or LM3 cells with depletion of endogenous PFKL. h, LM3 cells with depleted endogenous PFKL and reconstituted expression of WT Flag-rPFKL or Flag-rPFKL T331A were treated with or without SB203580 (10 μM) for 30 min before 40 min of glucose deprivation. Immunoprecipitation analyses with an anti-Flag antibody were performed. i, LM3 cells with depleted endogenous PFKL and reconstituted expression of WT Flag-rPFKL or Flag-rPFKL T331A were treated with or without 40 min of glucose deprivation. Immunoprecipitation analyses with an anti-Flag antibody were performed.
Extended Data Fig. 3 PFKL acts as a protein kinase and phosphorylates PLIN2 S159.
f, l, r, s, Immunoblotting analyses with the indicated antibodies were performed, presenting representative results from three independent experiments. a, b, The crystal structures of PFKP dimer (PDB: 4XYJ) (a) and PFKL dimer generated by homology modeling based on PFKP (b). c, Molecular dynamics simulations of the nonphosphorylated PFKL and the T331-phosphorylated PFKL dimers were performed. d, Coomassie blue staining of purified tag-free PFKL proteins. e, The activity of the WT PFKL (mock-phosphorylated control), WT PFKL (pT331), PFKL T331A and T331D mutants with respect to F6P titrations (ATP 0.5 mM) or ATP titrations (F6P 4 mM) is presented. Two preparations of protein were used. f, Flag-rPFKL proteins were expressed in Huh7 cells with depletion of endogenous PFKL (left). The media were collected for analysis of remaining glucose (middle) or lactate production. n = 3. The P values were labeled in front of the corresponding data points (right). g-k, Huh7 cells with depleted endogenous PFKL and reconstituted expression of Flag-rPFKL proteins were subjected to ECAR analysis by Seahorse; n = 8 (g), glycolytic metabolites analysis by LC-MS/MS; n = 3 (h), metabolic flux analysis by LC-MS/MS; n = 3 (i, k), glycogen level analysis; n = 3 (j). l, An in vitro kinase assay was performed by mixing Flag-PFKL with His-PLIN2 in the presence of [γ-32P]ATP. Autoradiography and immunoblotting analyses were performed. m, The binding ability of ATP to His-SUMO-PFKL proteins was measured by biolayer interferometry (Sartorius Octet) assay. Representative results from three independent experiments are presented. n, Enzyme kinetics plots of velocity relative to PLIN2 concentrations between PFKL WT and T331D. The Vmax and Km of PFKL in phosphorylating PLIN2 were calculated. n = 3. o, Stoichiometry of PLIN2 phosphorylation by PFKL T331D was presented. n = 3. p, An in vitro phosphorylation assay was performed by mixing His-PLIN2 with Flag-PFKL. LC-MS/MS analyses showed a tryptic fragment at m/z 805.85974 Da (-6.52 mmu/-8.09 ppm), which was matched with the +4 charged peptide 154-SVVSGp(S)INTVLGSRMMQL VSSGVENALTK-182 of PLIN2, suggesting that PLIN2 S159 was phosphorylated. The XCorr score was 2.8. q, Alignment of protein sequences spanning PLIN2 S159 in different species (top). The human PLIN2 S159 peptide sequence and the corresponding sequence of PLIN3 were aligned (bottom). r, WT Flag-rPLIN2 and Flag-rPLIN2 S159A were expressed in Huh7 or LM3 cells with depletion of endogenous of PLIN2 (top). PLIN2 depletion efficiency was quantified. n = 3 (bottom). s, Huh7 cells stably expressing shCtrl or shPLIN2 were treated with 40 min of glucose deprivation. t, IHC analyses of human HCC samples were performed with the indicated antibodies in the presence or absence of a blocking peptide for PLIN2 pS159. Representative images from three independent experiments are shown. u, Molecular docking analyses of F6P in the catalytic domain of nonphosphorylated PFKL (top) or T331-phosphorylated PFKL (bottom). e-k, n, o, r, Data are presented as means ± SD of three biologically independent replicates, analyzed by two-sided unpaired Student’s t-test (h, r), one-way ANOVA (f, g) or two-way ANOVA (i, k).
Extended Data Fig. 4 PFKL-mediated PLIN2 S159 phosphorylation promotes the interaction between PLIN2 and CPT1A and lipid droplet-mitochondria tethering.
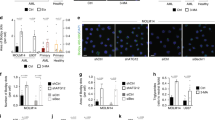
a, c, Immunoblotting analyses with the indicated antibodies were performed, presenting representative results from three independent experiments. d, f, g, Representative images from three independent experiments. a, LM3 cells with depleted endogenous PLIN2 and reconstituted expression of Flag-rPLIN2 proteins were treated with 40 min of glucose deprivation. b, A schematic depicting Red C12 transport assay. c, d, e, Flag-rPLIN2proteins were expressed in Huh7 cells with depletion of endogenous PLIN2. Immunoprecipitation analyses with an anti-Flag antibody were performed (c). Cells were labeled with GFP-LiveDrop and mito-DsRed. Percentages of mitochondria-associated LDs per cell were quantified. n (WT) = 32, n (S159D) = 36 (d). LDs number and the percentage of LDs area per cell were quantified. n (WT) = 55, n (S159D) = 56 (e). f, Huh7 cells with depleted endogenous PFKL and reconstituted expression of Flag-rPFKL proteins were stimulated by 40 min of glucose deprivation (left). Percentages of mitochondria-associated LDs per cell were quantified. n (WT) = 26, n (T331A) = 28. g-i, Huh7 cells with depleted endogenous PFKL and reconstituted expression of Flag-rPFKL proteins were incubated with Red C12 for 16 h before treatment with 4 h of glucose deprivation (g). Mitochondrial translocation of Red-C12 from LDs upon glucose starvation was quantified. n (WT) = 30, n (T331A) = 30. A.U., arbitrary unit (h). LDs number and the percentage of LDs area per cell were quantified. n (Glc+) = 30, n (Glc‒) = 30 (i). d-f, h, i, Data are presented as means ± SD of three biologically independent replicates, analyzed by two-sided unpaired Student’s t-test (d-f, h) or two-way ANOVA (i).
Extended Data Fig. 5 PLIN2 S159 phosphorylation-mediated lipid droplet consumption is independent of autophagy.
a, f, g, i, j, Immunoblotting analyses with the indicated antibodies were performed, presenting representative results from three independent experiments. b-d, e, k, Representative images from three independent experiments. a, Huh7 cells were infected with lentiviruses expressing shCtrl or shATG5. b, Huh7 cells stably expressing shCtrl or shATG5 were treated with 4 h of glucose deprivation (left). LDs number and the percentage of LDs area per cell were quantified. n (shCtrl) = 59, n (shATG5) = 58 (right). c, d, Huh7 cells with depleted endogenous PFKL (c) or PLIN2 (d) and reconstituted expression of Flag-rPFKL (c) or Flag-rPLIN2 (d) proteins were infected with lentiviruses expressing shATG5. Cells were treated with 4 h of glucose deprivation. LDs number and the percentage of LDs area per cell were quantified. n (shATG5/PFKL + rPFKL WT, Glc+) = 52, n (shATG5/PFKL + rPFKL WT, Glc‒) = 53, n (shATG5/PFKL + rPFKL T331A, Glc+) = 56, n (shATG5/PFKL + rPFKL T331A, Glc‒) = 51 (c). n (shATG5/PLIN2 + rPLIN2 WT, Glc+) = 60, n (shATG5/PLIN2 + rPLIN2 WT, Glc‒) = 50, n (shATG5/PLIN2 + rPLIN2 S159A, Glc+) = 61, n (shATG5/PLIN2 + rPLIN2 S159A, Glc‒) = 59 (d). e, Workflow for the ATGL recruitment assay (left). The LDs were stained with LipidTOX Deep Red and immunofluorescence analyses with an anti-ATGL antibody were performed (right). f, The precipitated LDs that were incubated with His-SUMO-ATGL as described in (e) were subjected to immunoblotting. g, WT Flag-rPLIN2, Flag-rPLIN2 S159D or a Flag-Vector was expressed in Huh7 cells with depletion of endogenous PLIN2. Immunoprecipitation analyses with an anti-Flag antibody were performed. h, Molecular dynamics simulations of the WT PLIN2 and PLIN2 S159D were performed. i, PLIN2 proteins immunoprecipitated from endogenous PLIN2-depleted Huh7 cells with reconstituted expression of WT rPLIN2 or rPLIN2 S159D were digested with 2.5 ng/mL trypsin for 5 min at room temperature. j, Huh7 cells were infected with lentiviruses expressing shCtrl or shATGL. k, Huh7 cells stably expressing shCtrl or shATGL were stimulated by 40 min of glucose deprivation (left). Percentages of mitochondria-associated LDs per cell were quantified. n (shCtrl) = 25 and n (shATGL) = 26. l, m, Seahorse FAO assays were performed in Huh7 cells with depleted endogenous PLIN2 (l) or PFKL (m) and reconstituted expression of the indicated Flag-rPLIN2 or rPFKL proteins with or without 2-DG treatment. n = 4. The P values were labeled in front of the corresponding data points. P < 0.0001 (orange) for the difference between WT rPLIN2 (l) or rPFKL (m) 2-DG+ (orange) and WT rPLIN2 (l) or rPFKL (m) 2-DG‒ (blue); P < 0.0001 (magenta) for the difference between rPLIN2 S159A (l) or rPFKL T331A (l) 2-DG+ (magenta) and WT rPLIN2 (m) or rPFKL (l) 2-DG+ (orange). n, o, Seahorse FAO assays were performed in Huh7 cells (n) or Huh7 cells with depleted endogenous PLIN2 and reconstituted expression of Flag-rPLIN2 S159D (o) treated with or without Etomoxir (Eto, 40 μM) before treatment with or without glucose deprivation. n = 8. The P values were labeled in front of the corresponding data points. P < 0.0001 (orange) for the difference between Huh7 Glc‒ (orange) and Huh7 Glc‒ (pretreated with Eto, magenta). P < 0.0001 (green) for the difference between rPLIN2 S159D Glc+ (green) and rPLIN2 S159D Glc+ (pretreated with Eto, red). p, Huh7 cells with depleted endogenous PFKL and reconstituted expression of Flag-rPFKL proteins were subjected to LC-MS/MS. n = 3. q, Huh7 cells with depleted endogenous PFKL or PLIN2 and reconstituted expression of the indicated Flag-rPFKL proteins or Flag-rPLIN2 proteins were treated with 2 h of glucose deprivation (no FBS). The levels of cellular ATP were examined. n = 3. b-d, k-q, Data are presented as means ± SD of three biologically independent replicates, analyzed by two-sided unpaired Student’s t-test (k), one-way ANOVA (l-o), two-way ANOVA (b-d, p, q).
Extended Data Fig. 6 PFKL-mediated PLIN2 S159 phosphorylation promotes the survival and proliferation of HCC cells and tumor growth.
a, Huh7 cells (2 × 104 cells) with depleted endogenous PLIN2 or PFKL and reconstituted expression of the indicated Flag-rPLIN2 (left) or Flag-rPFKL (right) were treated with or without 2-DG (25 mM) or Etomoxir (Eto, 40 μM) for 48 h. The cells were counted. Data are presented as means ± SD of three biologically independent replicates, analyzed by one-way ANOVA; n = 5. b-e, Huh7 cells with depleted endogenous PLIN2 or PFKL and reconstituted expression of the indicated Flag-rPLIN2 or Flag-rPFKL were subcutaneously injected into six-week-old male athymic BALB/c nude mice (n = 12 per group). Ten days after tumor cell injection, 0.2 mL of 2-DG (500 mg/kg) or PBS control was intraperitoneally injected daily for 18 days (n = 6). The tumor volumes (b) and weights (c) were determined. Data are presented as means ± SD of 6 biologically independent replicates, analyzed by one-way ANOVA. The indicated tumor tissues were analyzed by TUNEL assay. Representative images from three independent experiments are shown. Apoptotic cells in 10 microscope fields were quantified. Data were analyzed by two-sided unpaired Student’s t-test; n = 10 (d, e).
Extended Data Fig. 7 PLIN2 S159 is phosphorylated and upregulated in human liver cancer.
Analysis of the dataset from the Cancer Proteogenomic Data Analysis Site (https://cprosite.ccr.cancer.gov/) was performed. Adjacent normal samples: minimum, ‒1.688; maximum, ‒0.0815; lower quartile, ‒1.1200; upper quartile, ‒0.5775; lower fence, ‒1.1688; upper fence, ‒0.0815; and median, ‒0.9422; Tumor samples: minimum, ‒1.6302; maximum, 1.9100; lower quartile, ‒0.7998; upper lower quartile, 1.3789; lower fence, ‒1.6302; upper fence, 1.9100; and median, 0.4399. The horizontal lines mark the median Data are presented as the means ± SD. Data were analyzed by two-sided unpaired Student’s t-test; n = 15 (Adjacent normal samples) and n = 13 (Tumor samples).
Supplementary information
Supplementary Information
Supplementary methods
Source data
Source Data Fig. 1
Unprocessed western blots
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Unprocessed western blots
Source Data Fig. 3
Unprocessed western blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed western blots
Source Data Extended Data Fig./Table 1
Statistical Source Data
Source Data Extended Data Fig. 2
Unprocessed western blots
Source Data Extended Data Fig. 3
Unprocessed western blots
Source Data Extended Data Fig./Table 3
Statistical Source Data
Source Data Extended Data Fig. 4
Unprocessed western blots
Source Data Extended Data Fig./Table 4
Statistical Source Data
Source Data Extended Data Fig. 5
Unprocessed western blots
Source Data Extended Data Fig./Table 5
Statistical Source Data
Source Data Extended Data Fig./Table 6
Statistical Source Data
Source Data Extended Data Fig./Table 7
Statistical Source Data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meng, Y., Guo, D., Lin, L. et al. Glycolytic enzyme PFKL governs lipolysis by promoting lipid droplet–mitochondria tethering to enhance β-oxidation and tumor cell proliferation. Nat Metab (2024). https://doi.org/10.1038/s42255-024-01047-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42255-024-01047-2