Abstract
In our previous study, we developed a triple-negative breast cancer (TNBC) subtype classification that correlated with the TNBC molecular subclassification. In this study, we aimed to evaluate the predictor variables of this subtype classification on the whole slide and to validate the model’s performance by using an external test set. We explored the characteristics of this subtype classification and investigated genomic alterations, including genomic scar signature scores. First, TNBC was classified into the luminal androgen receptor (LAR) and non-luminal androgen receptor (non-LAR) subtypes based on the AR Allred score (≥ 6 and < 6, respectively). Then, the non-LAR subtype was further classified into the lymphocyte-predominant (LP), lymphocyte-intermediate (LI), and lymphocyte-depleted (LD) groups based on stromal tumor-infiltrating lymphocytes (TILs) (< 20%, > 20% but < 60%, and ≥ 60%, respectively). This classification showed fair agreement with the molecular classification in the test set. The LAR subtype was characterized by a high rate of PIK3CA mutation, CD274 (encodes PD-L1) and PDCD1LG2 (encodes PD-L2) deletion, and a low homologous recombination deficiency (HRD) score. The non-LAR LD TIL group was characterized by a high frequency of NOTCH2 and MYC amplification and a high HRD score.
Similar content being viewed by others
Introduction
Breast cancers that do not express the estrogen receptor (ER) and the progesterone receptor (PR) and that do not overexpress human epidermal growth factor receptor 2 (HER2) are collectively grouped as triple-negative breast cancer (TNBC). This subtype accounts for 15–20% of primary breast cancers1 and exhibits more aggressive clinical behavior than other breast cancer subtypes2,3. Despite being categorized as a single disease, TNBC is highly heterogeneous4.
Researchers at Vanderbilt University, analyzed the gene expression profile of 587 TNBC cases and classified TNBC into six subtypes (the TNBCtype-6 classification)5. Basal-like 1 (BL1) is enriched for cell cycle and DNA damage response genes. Basal-like 2 (BL2) is characterized by enrichment in growth factor signaling. Immunomodulatory (IM) is defined by high expression of immune-related pathways. Mesenchymal (M) is characterized by genes related to mesenchymal differentiation and proliferation. Mesenchymal stem–like (MSL) has mesenchymal features but low levels of proliferation. Luminal androgen receptor (LAR) is defined by the activation of hormone-related pathways, specifically through the androgen receptor (AR). Later, the same group refined the molecular subclassification into four subtypes (BL1, BL2, M, and LAR; the TNBCtype-4 classification). This refinement was based on the observation of the significant influence of tumor-infiltrating lymphocytes (TILs) in the IM subtype and stromal cells in the MSL subtype6.
In our previous study, we developed a TNBC subtype classification that correlated with the Vanderbilt TNBCtype gene expression molecular subclassification, making TNBC subtyping more widely applicable7. We analyzed the gene expression profile of 145 TNBC cases and subtyped them into the re-classified LAR, IM, BL1, and M Vanderbilt TNBC subtypes. We used a classification and regression tree (CART) prediction model for this subtype classification. The predictor variables adopted for the subtype classification in the model were the TIL score and immunohistochemistry (IHC) of the AR and p16, which are commonly used in pathology laboratories. The concordance between this subtype classification and the Vanderbilt subtypes was 0.71. However, we conducted this previous study on a tissue microarray (TMA) for IHC and TIL scores, and it lacked independent external validation. A TMA includes small tissue cores from various regions of the tumor, but it only provides a limited representation of the tumor. By contrast, whole-slide analysis allows for a comprehensive evaluation of the entire tumor and encompasses its full heterogeneity. This approach provides an accurate representation of tumor biology and helps to identify crucial molecular features for subtype classification and characterization. Studies without external validation may encounter several problems, including a lack of generalizability to different datasets, a risk of overestimation of the results, and an inability to assess model performance accurately. Therefore, our proposed TNBC subtype classification requires additional assessment using whole-slide analysis for IHC and external validation to ensure its robustness.
The homologous recombination DNA repair (HRR) pathway is involved in the repair of DNA double-strand breaks8. The genomic scar signature score is associated with BRCA1 and BRCA2 germline or somatic mutations, BRCA1 and RAD51C promoter methylation, and PALAB2 germline mutation in TNBC9,10. In addition to germline BRCA1 and BRCA2 mutations, which affect a key gene in the HRR pathway11, somatic or epigenetic BRCA1 and BRCA2 alterations, defects in other HRR-modulating genes, and high genomic scar signature scores are emerging as potential predictive markers showing sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitors12. The BL1 and BL2 subtypes of the Vanderbilt TNBCtype classification display enrichment of genes associated with cell proliferation and the DNA damage response. In almost all cell lines with mutations in BRCA1 and BRCA2, gene expression is correlated with the basal-like subtype5, suggesting that DNA repair–targeting agents are a suitable treatment.
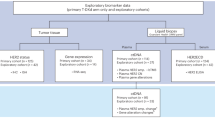
In this study, we evaluated the predictor variables of our previous TNBC subtype classification, p16 and AR expression and TIL scores, on whole slides, which are used in pathology laboratories, rather than TMAs to capture the overall tumor biology and to improve subtype classification. In addition, we sought to validate the model’s performance by using an external test set to assess its robustness. We then explored the characteristics of this subtype classification and investigated genomic alterations, including genomic scar signature scores. The study design is shown in Fig. 1.
Study design. TNBC, triple-negative breast cancer; TILs, tumor-infiltrating lymphocytes; AR, androgen receptor; IHC, immunohistochemistry; TCGA, The Cancer Genome Atlas; LAR, luminal androgen receptor; IM, immunomodulatory; BL1, basal-like 1; M, mesenchymal; LP, lymphocyte-predominant; LI, lymphocyte-intermediate; LD, lymphocyte-depleted. * Instead of the AR expression results in the TCGA dataset, the Vanderbilt classification based on gene expression was used for the LAR subtypes.
Results
Clinical characteristics and gene expression-based TNBC molecular subclassification of the training and test sets
The training set had a significantly higher frequency of metaplastic carcinoma and a significantly higher histological grade than the test set. Age, tumor size, and the American Joint Committee on Cancer (AJCC) pathologic stage were not significantly different between the training and test sets (Supplementary Table S1).
According to the TNBC molecular subclassification, 142 TNBC cases in the training set and 175 TNBC cases in the test set were assigned to one of the LAR, IM, BL1, or M subtypes. In the training set, there were 21 cases (14.8%) of the LAR subtype, 31 cases (21.8%) of the IM subtype, 25 cases (17.6%) of the BL1 subtype, and 38 cases (26.8%) of the M subtype. In the test set, there were 24 cases (13.7%) of the LAR subtype, 31 cases (17.7%) of the IM subtype, 37 cases (21.1%) of the BL1 subtype, and 42 cases (24.0%) of the M subtype. There were 27 unclassified cases (19.9%) in the training set and 41 unclassified cases (23.4%) in the test set; we excluded these cases. There was no significant difference in the TNBC molecular subclassification between the two sets (p = 0.700) (Supplementary Table S1).
Subtype classification of TNBC into LAR subtype and TIL groups
The CART prediction model created a subtype classification that corresponds to the TNBC molecular subclassification. We selected the AR Allred score and the TIL score as markers for this subtype classification, but we excluded the p16 staining pattern. The CART prediction model selected a cut-off of 6 points for the AR Allred score and 20% and 60% for the TIL score. Based on the prediction of the CART model, this subtype classification first classified TNBC into two subtypes according to the AR Allred score: the LAR subtype for a score ≥ 6, and the non-LAR subtype for a score < 6. Next, the non-LAR subtype was further subdivided into three TIL groups with the TIL score cut-offs of 20% and 60%: the lymphocyte-predominant (LP), lymphocyte-intermediate (LI), and lymphocyte-depleted (LD) groups. The subtype classification was as follows: LAR subtype, AR Allred score ≥ 6; non-LAR subtype LP group (abbreviated as the LP group), AR Allred score < 6 and TIL score ≥ 60%; non-LAR subtype LI group (abbreviated as the LI group), AR Allred score < 6 and 20% ≤ TIL score < 60%; non-LAR subtype LD group (abbreviated as the LD group), AR Allred score < 6 and TIL score < 20% (Fig. 2). The LAR subtype corresponded to the LAR molecular subtype. Among the non-LAR subtypes, the LP group corresponded to the IM molecular subtype, the LI group to the BL1 molecular subtype, and the LD group to the M molecular subtype.
Triple-negative breast cancer (TNBC) subtype classification and performance metrics. (a) Decision tree for classifying the TNBC subtypes based on the AR Allred score and percentage of stromal TILs, dividing cases into the LAR, LP, LI, and LD subtypes. (b) Performance of the classification and regression tree prediction model in the training and test sets, evaluated with the accuracy metrics recall, precision, balanced accuracy, the F1 score, overall accuracy, and the kappa statistic. AR, androgen receptor; LAR, luminal androgen receptor; TILs, tumor-infiltrating lymphocytes; LP, lymphocyte-predominant; LI, lymphocyte-intermediate; LD, lymphocyte-depleted.
F1 score and concordance between the subtype classification and the TNBC molecular subclassification
The overall concordance between the subtype classification and the TNBC molecular subclassification was 0.66 (95% confidence interval [CI] 0.57–0.75; κ = 0.54) in the training set and 0.56 (95% CI 0.47–0.65; κ = 0.40) in the test set. In both sets, the LP group showed the highest precision (also called the positive predictive value), while the LI group showed the lowest precision among the four subtypes. The LAR subtype had the highest recall (referred to as sensitivity), while the LI group had the lowest, similarly to its precision. The F1 score of the LI group was the lowest, reflecting its precision and recall values (Fig. 2b).
The differences in clinicopathologic characteristics among the LAR subtype and the TIL groups
Among 317 cases, including those in the training and test sets, the LAR subtype comprised 71 cases (22.3%), the LP group included 73 cases (23.0%), the LI group included 59 cases (18.6%), and the LD group included 114 cases (36.0%). The age at diagnosis, histologic type, and tumor size showed significant differences among the LAR subtype and the TIL groups. The LAR subtype was associated with the oldest age. Although the number was small, all four cases of invasive lobular carcinoma had the LAR subtype, and all four cases of salivary gland–like carcinoma were in the LD group. Carcinoma with medullary features was the most frequent in the LP group, and metaplastic carcinoma was the most common in the LD group. The LAR subtype showed a lower histologic grade than the non-LAR subtype. The LP group had the smallest tumor size. There was no significant difference in the AJCC pathologic stage among the subtypes (Table 1).
Log-rank test and multivariable cox regression analysis in the LAR subtype and the TIL groups
The log-rank test showed significant differences in survival outcomes among the subtypes for overall survival (OS) (p = 0.003), relapse-free survival (RFS) (p < 0.001), and distant metastasis–free survival (DFS) (p = 0.004) (Fig. 2). In the multivariable survival analysis, the LD group was an independent prognostic factor for poor OS, RFS, and DFS. The LD group had significantly worse OS than the LP group (hazard ratio [HR] 11.8, 95% CI 1.55–89.7, p = 0.017). The risk of recurrence was significantly higher in the LD group than in the LP group (HR 3.68, 95% CI 1.70–7.93, p < 0.001). The risk of metastasis was significantly higher in the LD group than in the LP group (HR 3.81, 95% CI 1.67–8.67, p = 0.001) (Fig. 3). In separate analyses of the training and test sets, the LD subtype was associated with significantly worse prognosis for OS, RFS, and DFS in the training set. This association was not found to be statistically significant in the test set (Supplementary Fig. 1).
Survival analysis. Kaplan–Meier curves and the forest plots for multivariable Cox proportional hazards regression for overall survival (a, b), relapse-free survival (c, d), and distant metastasis-free survival (e, f) according to the triple-negative breast cancer (TNBC) subtype classification. LAR, luminal androgen receptor; TILs, tumor-infiltrating lymphocytes; LP, lymphocyte-predominant; LI, lymphocyte-intermediate; LD, lymphocyte-depleted.
Hallmark signatures of the LAR subtype and the TIL groups
Of the 50 hallmark signatures, 31 were significantly different among the LAR subtype and the TIL groups. Eleven hallmark signatures had an adjusted p < 1 × 10−5. The LAR subtype showed a significantly higher gene set enrichment analysis (GSEA) score than the TIL groups in metabolism hallmark signatures, including bile acid metabolism, fatty acid metabolism, and xenobiotic metabolism. The LP group showed a significantly higher GSEA score than the other TIL groups and the LAR subtype in immune hallmark signatures, including allograft rejection, IL6 JAK STAT3 signaling, the interferon-alpha response, and the interferon-gamma response. The LD group showed a significantly higher GSEA score for epithelial–mesenchymal transition, hypoxia, myogenesis, and transforming growth factor (TGF)-beta. The GSEA scores are presented graphically in Fig. 4. The 11 hallmark signatures with an adjusted p < 1 × 10−5 are summarized in Fig. 5.
Hallmark signatures and differences according to the triple-negative breast cancer (TNBC) subtype classification. Each cell in the heatmap represents the gene signature value. Each column represents samples grouped by subtype classification. Each row represents a hallmark signature grouped by category. The left column with annotation represents the minus logarithm (base 10) of the adjusted p-values. This heatmap is generated using the tidyHeatmap R package (version 1.8.1). LAR, luminal androgen receptor; LP, lymphocyte-predominant; LI, lymphocyte-intermediate; LD, lymphocyte-depleted; CC, cellular component; DD, DNA damage; DV, development; PR, proliferation.
Violin plots of significantly different hallmark signatures according to the triple-negative breast cancer (TNBC) subtype classification. Post hoc analysis of hallmark signatures with an adjusted p < 1 × 10−5 was performed. The first row of signatures is related to metabolism; the second to immunity; and the third to epithelial-mesenchymal transition, hypoxia, myogenesis, and transforming growth factor-β. LAR, luminal androgen receptor; LP, lymphocyte-predominant; LI, lymphocyte-intermediate; LD, lymphocyte-depleted; ns, p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p = 0.0001.
Genomic alterations in the LAR subtype and the TIL groups using The Cancer Genome Atlas (TCGA) dataset
From the TNBC molecular subclassification evaluated by Bareche et al.13 and whole hematoxylin and eosin (H&E) slide images from TCGA, 148 TNBC cases were available for subtype classification into the LAR subtype and the TIL groups in the firehose dataset, and 76 cases were available in the TCGA invasive breast cancer/Nature 2012 dataset. In the firehose dataset, there were 26 cases (16.1%) in the LAR subtype, 21 cases (13.0%) in the LP group, 34 cases (21.1%) in the LI group, and 80 cases (49.7%) in the LD group. In the TCGA invasive breast cancer/Nature 2012 dataset, there were 9 cases (11.8%) in the LAR subtype, 14 cases (18.4%) in the LP group, 15 cases (19.7%) in the LI group, and 38 cases (50.5%) in the LD group.
The LAR subtype was characterized by a high PIK3CA mutation rate, high CD274 (encodes PD-L1) and PDCD1LG2 (encodes PD-L2) deletion, and a low genomic scar signature score. The LD group was characterized by a high NOTCH2 and MYC amplification rate and a high genomic scar signature score. The non-LAR subtypes were characterized by a low CD274 and PDCD1LG2 deletion rate and a high B2M deletion rate compared with the LAR subtype. The frequency of BRCA1 and BRCA2 germline and somatic mutations did not show significant differences among the LAR subtype and the TIL groups. The genomic scar signature score was significantly higher in the LI and LD groups than in the LAR subtype (Fig. 6).
Genomic analysis of the triple-negative breast cancer (TNBC) subtype classification in The Cancer Genome Atlas (TCGA) dataset. (a) Oncoplot visualizing the frequency of molecular alterations across samples, categorized by the TNBC subtype (LAR, LP, LI, LD). Alterations are depicted by color-coded mutation types. The bar plot on the right indicates the total number of alterations per sample. This oncoplot is generated using the maftools R package (version 2.16.0). (b) Bar plots of significantly different genomic alterations. (c) Violin plot for the genomic scar signature. The difference in genomic scar signature scores was statistically evaluated using the Kruskal–Wallis test followed by the post hoc Wilcoxon signed-rank test. LAR, luminal androgen receptor; LP, lymphocyte-predominant; LI, lymphocyte-intermediate; LD, lymphocyte-depleted.
Discussion
In this study, we attempted to classify TNBC into subtypes that correlate with the TNBC molecular subclassification by using whole-slide images to evaluate TIL scores and IHC of p16 and AR, which were selected as predictor variables from the previous subtype classification. The CART prediction model selected the AR Allred score (cut-off: 6) and the TIL score (cut-off: 20% and 60%) as the predictor variables, but not the p16 staining pattern. TNBC was categorized into LAR and non-LAR subtypes, and the non-LAR subtype was further divided into the TIL groups. The overall accuracy of subtype classification into the TNBC molecular subclassification was 0.66 (κ = 0.54) in the training set and 0.56 (κ = 0.40) in the test set. Although this subtype classification showed low agreement with the TNBC molecular subclassification, each of the four subtypes in this classification displayed specific characteristics. The distinct clinicopathologic and genomic features of the proposed TNBC subtype classification is summarized in Table 2. Our proposed AR Allred score and TIL score cut-offs are supported by evidence from clinical and genetic analyses, which have been used arbitrarily in previous studies. In addition, our findings suggest that only AR IHC and the TIL scores could be used to subtype TNBC, indicating the potential for a clinically feasible TNBC stratification approach.
AR is expressed in approximately 10–43% of TNBC cases, depending on the definition of AR positivity used. Some studies used a threshold of more than 10% of tumor cells showing AR staining14, while others used an Allred score of ≥ 3, similar to the criteria used for estrogen receptor (ER) and progesterone receptor (PR) positivity15. The H-score method has also been used to determine positive AR expression16. These variations in the definition of positive AR expression may lead to confusion, and the prevalence rates of AR-positive TNBC cases may vary among studies due to the use of different criteria. Positive criteria for ER and PR expression have been crucial to classify breast cancer and to guide hormonal therapy decisions, and substantial research and effort have been dedicated to establishing the current criteria. Likewise, the establishment of the criteria for defining positive AR expression is also necessary for the accurate classification of TNBC. In this study, based on TNBC molecular subclassification, we proposed a criterion for AR expression that allowed the division of TNBC cases into the LAR and non-LAR subtypes. The LAR subtype showed distinct characteristics, such as diagnosis at an older age and a lower histologic grade compared with the non-LAR subtype. This subtype displayed hallmark signatures associated with metabolism, a high rate of PIK3CA mutation and CD274 and PDCD1LG2 deletion, and a low genomic scar signature score. Similarly to our study, previous research has shown that the LAR subtype is associated with a high frequency of older age and invasive lobular carcinoma17, and enriched gene signatures are associated with hormone-related gene signaling pathways (including steroid hormone, porphyrin metabolism, and androgen/estrogen metabolism) and a high frequency of PIK3CA mutations5. Studies have reported that patients with AR-positive TNBC showed better DFS18 and a more favorable response to neoadjuvant chemotherapy19. We observed that the LAR subtype also showed a favorable prognosis in terms of OS, RFS, and DFS. Hence, it is possible that an Allred score of 6, as determined by molecular subclassification, could serve as a potential cut-off for the LAR subtype.
Recently, AR-negative TNBC, which accounts for approximately 67%–90% of TNBC cases, has been considered a separate molecular subtype from AR-positive TNBC and is referred to as quadruple-negative breast cancer (QNBC)20,21. QNBC does not benefit from AR antagonists. Studies suggesting a worse prognosis for QNBC22 compared with AR-positive TNBC indicate the need to understand the biological basis and to explore alternative therapeutic strategies for QNBC. Recent research has uncovered fatty acyl-CoA synthetase 4, S-phase kinase associated protein 2, EGFR, and CD151 as potential candidates20. As mentioned above, the variability in the definition of AR positivity complicates the study of QNBC. The non-LAR subtype proposed in our study, based on gene expression results and with evidence of distinctive features, could be used as a surrogate for QNBC. The poorer prognosis of the non-LAR subtype compared with the LAR subtype is consistent with QNBC. The non-LAR subtype may help to define QNBC in future studies.
Studies have reported that high TILs can have predictive value for the treatment efficacy of immune checkpoint inhibitors23 or neoadjuvant chemotherapy24, and are related to a better response, a pathologic complete response (pCR), and survival25,26. According to the 2014 International TILs Working Group27, the level of TILs is assessed using 10% increments. However, it appears that stratification of TIL levels is necessary in practice. Categorizing TILs into groups, such as low, intermediate, and high, improves the interpretability of the results, supports clinical decision making, and allows for better comparison and standardization across studies. This approach can facilitate clinical translation, making it easier for researchers and clinicians to analyze and communicate the significance of TILs in different contexts. In this study, we divided TILs into three groups based on molecular subclassification, each with distinct characteristics. The LP group had a high frequency of carcinoma with medullary features and the smallest tumor size, immune hallmark signatures, and good OS. The LD group showed a high frequency of metaplastic carcinoma and salivary gland-like carcinoma; significantly poorer OS, RFS, and DFS; the mutational signature of epithelial–mesenchymal transition, hypoxia, myogenesis, and TGF-beta; a high rate of NOTCH2 and MYC amplification; and a high genomic scar signature score. The identification of distinct characteristics and clinical outcomes associated with each TIL group suggests that subdivision into TIL groups with 20% and 60% cut-offs for the TIL score may be valid.
In our genomic analysis using the TCGA dataset, the genomic scar signature score was higher in the LI and LD groups than in the LAR subtype or the LP group. Our subtype classification suggests that high genomic scar signature scores are related to the non-LAR subtype and lower TILs. This result suggests that our subtype classification could potentially be applied to stratify patients with TNBC in clinical trials for treatment with PARP inhibitors.
A recent study28, which classified TNBC molecular subtypes by using gene expression microarrays, reported that the LAR molecular subtype showed favorable OS and DFS. The IM molecular subtype demonstrated relatively better DFS, while the M and BL2 molecular subtypes exhibited worse OS and DFS. Regarding the 3-year recurrence rate, the LAR molecular subtype showed a significantly better prognosis than the M and BL2 molecular subtypes. The results of our survival analysis were in close agreement to that study. In our study, the subtypes exhibited significantly different survival rates. The LAR subtype showed favorable OS, RFS, and DFS. For the non-LAR subtype, the LP group showed significantly better OS, RFS, and DFS compared with the LD group. These results support the idea that our subtype classification can serve as a prognostic marker. However, the lack of statistical significance in the test set suggests the need for further investigation. It highlights the need for additional studies with larger sample sizes to robustly validate the prognostic utility of our subtype classification.
This study has some limitations. There was relatively low agreement between our proposed TNBC subtype classification (LAR, LP, LI, and LD) and the re-classified Vanderbilt TNBCtype-4 classification (LAR, IM, BL1, and M), with an overall accuracy of 0.56 in the test set. The LI group contributed the most to lowering the overall accuracy among the four subtypes. The precision, recall, and F1 score were the lowest for BL1 molecular subtype classification in the LI group. The LI group did not show any distinct signature in our gene expression profile analysis, unlike the BL1 molecular subtype, which showed enrichment of genes involved in the cell cycle and DNA damage response (ATR/BRCA) pathway29,30. The LI group is probably different from the BL1 molecular subtype, and further research on this group is required.
TNBC clinically defined by ER, PR, and HER2 expression based on IHC differs from TNBC within the TCGA dataset defined by gene expression profiles. In addition, there were no AR expression results or AR scores based on IHC, so we could not divide the TNBC cases of the TCGA datasets into LAR and non-LAR subtypes by AR Allred score. So, we just defined the LAR molecular subtype of the Vanderbilt classification as the LAR subtype. To overcome this problem, we are conducting further research to characterize the genomic alterations of subtypes in clinically defined TNBC.
Several studies have attempted to classify TNBC subtypes using different IHC marker panels31,32,33. Although these studies have shown that TNBC classification was possible only with the IHC panel, they lacked comparisons with gene expression profiles. A study attempted IHC-based classification with a substantial agreement with messenger RNA (mRNA) expression–based FUSCC molecular subtyping34. This classification used AR, CD8, FOXC1, and DCL1 IHC to classify TNBC into the LAR, IM, BLIS, MES, and unclassified groups. This IHC-based classification was suggested as an independent prognostic factor for RFS in multivariable survival analysis. Indeed, this classification showed similarity to our subtype classification. However, our subtype classification is simpler, and our study evaluated genomic alterations in addition to gene expression signatures for each subtype.
In conclusion, we have presented a TNBC subtype classification that is easy to apply in pathology laboratories and correlates with molecular subclassification based on mRNA expression. Despite the low agreement with gene expression molecular subclassification, the TNBC subtype classification demonstrates distinct clinical and genomic characteristics. Our findings support a proposed AR Allred score cut-off of 6 for categorizing the LAR and non-LAR subtypes, and TIL score cut-offs of 20% and 60% for categorizing the TIL groups of the non-LAR subtype. These cut-offs appear to be useful for stratifying TNBC.
Methods
Dataset collection
The training set included the same 145 TNBC cases from Seoul St. Mary’s Hospital between January 2009 and October 2017 as in the previous study for the original subtype classification7. The test set comprised 191 cases of TNBC from Gangnam Severance Hospital from June 1997 to November 2014. After excluding the cases that failed the TIL score evaluation or IHC for AR and p16, the final training and test sets included 142 and 175 cases, respectively. The detailed inclusion and exclusion criteria were described in previous studies7,35. The need for informed consent was waived by the institutional review boards of Seoul St. Mary’s Hospital (KC21SISI0597) and Gangnam Severance Hospital (3-2013-0268).
RNA microarray and gene expression profile
RNA microarray analysis was performed to obtain gene expression data. Whole tissue and the same representative paraffin-fixed formalin-embedded tissue blocks employed for the microarray analysis were used. The Affymetrix Human Gene 2.0 ST Array and the Human Genome U133 Plus 2.0 Array were used in the training and test sets, respectively. The detailed protocols for total RNA isolation, RNA extraction, purification, labeling, hybridization, and the microarray assay were described in previous studies7,35. Raw gene expression profiles were normalized by quantile methods, log2 transformed, and centered around the median.
Gene expression-based TNBC molecular subclassification
Gene expression profiles from the microarray analysis of the training and test sets were uploaded to the TNBCtype website (http://cbc.mc.vanderbilt.edu/tnbc/) to classify TNBC into the Vanderbilt TNBCtype. For each case, correlation scores and corresponding p-values were obtained for each the six original subtypes (Vanderbilt TNBCtype: LAR, BL1, BL2, IM, M, and MSL)5.
Lehmann et al.5 refined the TNBC classification from six to four subtypes (LAR, BL1, BL2, and M) after observing the influence of TILs in the IM subtype and stromal cells in the MSL subtype. Bareche et al.13 observed that the BL2 and unstable (UNS) subtypes clustered non-specifically, indicating their lower reproducibility. However, given the importance of the immune response and the potential for immunotherapy in TNBC, it was necessary to maintain the IM subtype. Finally, we selected the LAR, IM, BL1, and M subtypes and named this re-classified Vanderbilt classification as the TNBC molecular subclassification. Then, we then proposed a subtype classification that could predict this TNBC molecular subclassification, as in the previous study. Each case was assigned to one of four subtypes with the highest correlation score and p < 0.05. Cases with p ≥ 0.05 for all four subtypes represented an unclassified group; we excluded these cases.
Evaluation of p16 and AR expression and TILs, which were selected as predictor variables in the previous subtype classification
P16 and AR expression and TIL scores, which were predictor variables in the previous subtype classification7, were evaluated in one whole representative section of each case from the training and test sets. An automatic IHC staining device (Ventana BenchMark ULTRA; Roche, USA) and antibodies targeting p16 (E6H4; Roche) and AR (SP107; Roche) were used. For p16, staining patterns were classified as negative, weak and mosaic, or diffuse and strong. AR was assessed as an Allred score, following the guidelines of St. Gallen and the American Society of Clinical Oncology/College of American Pathologists for ER and PR statining36,37 due to their similarity. The Allred score comprises a proportion score that reflects the percentage of AR-positive cells detected by immunohistochemistry (assigned 0–5 points for 0%, ≤ 1%, 1–10%, 11–33%, 34–66%, and ≥ 67%, respectively) and a nuclear intensity score (assigned 0, 1, 2, and 3 points for negative, weak, intermediate, and strong staining, respectively). Thus, the Allred score ranges from 0 to 8 points. The TIL score was determined by evaluating stromal TILs on the H&E-stained histology slides. The level of stromal TILs was assessed according to the 2014 International TILs Working Group and scored using 10% increments27.
CART prediction modeling
CART modeling was performed to predict the TNBC molecular subclassification using p16 and AR expression and TIL scores as predictor variables. The R package version rpart 4.1.19 was used for the modelling process. The parameters were set to 5 for maximum depth and 0.0001 for complexity. The subtype classification was developed based on the CART prediction model that correlates with the molecular subclassification. The performance of the subtype classification was evaluated in the training and test sets.
GSEA according to subtype classification
GSEA was performed on 336 cases, both the training and test sets, to investigate the associated pathways for each subtype38. The R package IOBR version 0.99.8 was used39. The difference in the signature score among the four subtypes was statistically evaluated with analysis of variance (ANOVA), and p-value adjustments were made using the false discovery rate (FDR). Post hoc analysis was conducted with Student’s t-test. The significance level was set at an adjusted p < 0.05.
Survival analysis according to subtype classification
The survival analysis was conducted on the entire dataset (n = 336), including the training and test sets, as well as separate analyses for each set. Kaplan–Meier survival curves and the log-rank test were used to compare the OS, RFS, and DFS among the subtypes. The Cox proportional-hazards test was performed for multivariable survival analysis, and the HR and its 95% CI were estimated. The significance level was set at p < 0.05.
Subtype classification of the TCGA dataset and genomic analysis
Genomic alterations of the subtype classification were investigated using genomic data from TCGA, namely the firehose dataset and the TCGA invasive breast cancer/Nature 2012 dataset downloaded from cBioPortal (https://www.cbioportal.org/). The firehose dataset was used to evaluate somatic mutations and copy number alterations. The TCGA invasive breast cancer/Nature 2012 dataset40 was used to evaluate BRCA1 and BRCA2 germline and somatic mutations.
The TNBC molecular subclassification in TCGA cases was previously evaluated by Bareche et al.13 and is available on the journal’s website. Therefore, TNBC cases within the TCGA dataset could be classified with the LAR subtype. The TIL score could be evaluated from the archived H&E-stained digital slide images (https://cancer.digitalslidearchive.org/).
Genomic alterations were analyzed according to subtype classification. Forty-two genes were selected according to a previous study by Lehmann et al.17. The frequency of mutations, gene deletions, and amplifications were analyzed according to subtype. Both homozygous and heterozygous deletions were considered gene deletions. Amplification, but not low level gain, was compared. The R package maftools version 2.16.0 was used to summarize and visualize mutations and copy number changes. Fisher's exact test and an FDR correction were applied. The significance level was set at an adjusted p < 0.05.
The genomic scar signature scores were estimated by Marquard et al.41 and downloaded from the journal’s website. The difference in genomic scar signature scores was statistically evaluated with the Kruskal–Wallis test followed by the post hoc Wilcoxon signed-rank test.
Ethics approval
The institutional review boards of Seoul St. Mary’s Hospital (KC21SISI0597) and Gangnam Severance hospital (3-2013-0268) approved this study. All procedures were performed in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Data availability
The datasets generated and/or analysed during the current study are available in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). The accession number for the training set data is GSE226289, and the accession numbers for the test set data are GSE157284 and GSE135565.
References
Criscitiello, C., Azim, H. A. Jr., Schouten, P. C., Linn, S. C. & Sotiriou, C. Understanding the biology of triple-negative breast cancer. Ann. Oncol. 23(Suppl 6), vi13–vi18. https://doi.org/10.1093/annonc/mds188 (2012).
Morris, G. J. et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer 110, 876–884. https://doi.org/10.1002/cncr.22836 (2007).
Haffty, B. G. et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 24, 5652–5657. https://doi.org/10.1200/jco.2006.06.5664 (2006).
Metzger-Filho, O. et al. Dissecting the heterogeneity of triple-negative breast cancer. J. Clin. Oncol. 30, 1879–1887. https://doi.org/10.1200/jco.2011.38.2010 (2012).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767. https://doi.org/10.1172/jci45014 (2011).
Lehmann, B. D. et al. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One 11, e0157368. https://doi.org/10.1371/journal.pone.0157368 (2016).
Yoo, T. K., Kang, J., Lee, A. & Chae, B. J. A triple-negative breast cancer surrogate subtype classification that correlates with gene expression subtypes. Breast Cancer Res. Treat. 191, 599–610. https://doi.org/10.1007/s10549-021-06437-8 (2022).
Bianchini, G., De Angelis, C., Licata, L. & Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 19, 91–113. https://doi.org/10.1038/s41571-021-00565-2 (2022).
Polak, P. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 49, 1476–1486. https://doi.org/10.1038/ng.3934 (2017).
Staaf, J. et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med. 25, 1526–1533. https://doi.org/10.1038/s41591-019-0582-4 (2019).
Glodzik, D. et al. Comprehensive molecular comparison of BRCA1 hypermethylated and BRCA1 mutated triple negative breast cancers. Nat. Commun. 11, 3747. https://doi.org/10.1038/s41467-020-17537-2 (2020).
Tung, N. M. et al. TBCRC 048: Phase II study of Olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J. Clin. Oncol. 38, 4274–4282. https://doi.org/10.1200/jco.20.02151 (2020).
Bareche, Y. et al. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 29, 895–902. https://doi.org/10.1093/annonc/mdy024 (2018).
Gucalp, A. et al. Phase II trial of bicalutamide in patients with androgen receptor–positive, estrogen receptor–negative metastatic breast cancer. Clin. Cancer Res. 19, 5505–5512 (2013).
Lee, E. G. et al. Androgen receptor as a predictive marker for pathologic complete response in hormone receptor–positive and HER-2-negative breast cancer with neoadjuvant chemotherapy. Cancer Res. Treat. 55, 542–550. https://doi.org/10.4143/crt.2022.834 (2023).
Bronte, G. et al. Androgen receptor expression in breast cancer: What differences between primary tumor and metastases?. Transl. Oncol. 11, 950–956. https://doi.org/10.1016/j.tranon.2018.05.006 (2018).
Lehmann, B. D. et al. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun. 12, 6276. https://doi.org/10.1038/s41467-021-26502-6 (2021).
Thike, A. A. et al. Loss of androgen receptor expression predicts early recurrence in triple-negative and basal-like breast cancer. Mod. Pathol. 27, 352–360. https://doi.org/10.1038/modpathol.2013.145 (2014).
Loibl, S. et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res. Treat. 130, 477–487. https://doi.org/10.1007/s10549-011-1715-8 (2011).
Hon, J. D. et al. Breast cancer molecular subtypes: From TNBC to QNBC. Am. J. Cancer Res. 6, 1864–1872 (2016).
Bhattarai, S., Saini, G., Gogineni, K. & Aneja, R. Quadruple-negative breast cancer: Novel implications for a new disease. Breast Cancer Res. 22, 127. https://doi.org/10.1186/s13058-020-01369-5 (2020).
He, J. et al. Prognostic value of androgen receptor expression in operable triple-negative breast cancer: A retrospective analysis based on a tissue microarray. Med. Oncol. 29, 406–410 (2012).
Loi, S. et al. LBA13—Relationship between tumor infiltrating lymphocyte (TIL) levels and response to pembrolizumab (pembro) in metastatic triple-negative breast cancer (mTNBC): Results from KEYNOTE-086. Ann. Oncol. 28, 608. https://doi.org/10.1093/annonc/mdx440.005 (2017).
Telli, M. L. et al. Association of tumor-infiltrating lymphocytes with homologous recombination deficiency and BRCA1/2 status in patients with early triple-negative breast cancer: A pooled analysis. Clin. Cancer Res. 26, 2704–2710. https://doi.org/10.1158/1078-0432.Ccr-19-0664 (2020).
Tan, Q. et al. Potential predictive and prognostic value of biomarkers related to immune checkpoint inhibitor therapy of triple-negative breast cancer. Front. Oncol. 12, 779786. https://doi.org/10.3389/fonc.2022.779786 (2022).
Echavarria, I. et al. Pathological response in a triple-negative breast cancer cohort treated with neoadjuvant carboplatin and docetaxel according to Lehmann’s refined classification. Clin. Cancer Res. 24, 1845–1852. https://doi.org/10.1158/1078-0432.Ccr-17-1912 (2018).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an international TILs working group 2014. Ann. Oncol. 26, 259–271. https://doi.org/10.1093/annonc/mdu450 (2015).
Masuda, H. et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 19, 5533–5540. https://doi.org/10.1158/1078-0432.Ccr-13-0799 (2013).
Lehmann, B. D. & Pietenpol, J. A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 232, 142–150. https://doi.org/10.1002/path.4280 (2014).
Yin, L., Duan, J. J., Bian, X. W. & Yu, S. C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22, 61. https://doi.org/10.1186/s13058-020-01296-5 (2020).
Choi, J., Jung, W. H. & Koo, J. S. Clinicopathologic features of molecular subtypes of triple negative breast cancer based on immunohistochemical markers. Histol. Histopathol. 27, 1481–1493. https://doi.org/10.14670/hh-27.1481 (2012).
Kim, S. et al. Feasibility of classification of triple negative breast cancer by immunohistochemical surrogate markers. Clin. Breast Cancer 18, e1123–e1132. https://doi.org/10.1016/j.clbc.2018.03.012 (2018).
Kumar, S. et al. Molecular subtyping of triple negative breast cancer by surrogate immunohistochemistry markers. Appl. Immunohistochem. Mol. Morphol. 29, 251–257. https://doi.org/10.1097/pai.0000000000000897 (2021).
Zhao, S. et al. Molecular subtyping of triple-negative breast cancers by immunohistochemistry: molecular basis and clinical relevance. Oncologist 25, e1481–e1491. https://doi.org/10.1634/theoncologist.2019-0982 (2020).
Ahn, S. G. et al. Clinical and genomic assessment of PD-L1 SP142 expression in triple-negative breast cancer. Breast Cancer Res. Treat 188, 165–178. https://doi.org/10.1007/s10549-021-06193-9 (2021).
Hammond, M. E. H. et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 134, e48–e72 (2010).
Fitzgibbons, P. L. et al. Template for reporting results of biomarker testing of specimens from patients with carcinoma of the breast. Arch. Pathol. Lab. Med. 138, 595–601. https://doi.org/10.5858/arpa.2013-0566-CP (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Zeng, D. et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol. 12, 687975. https://doi.org/10.3389/fimmu.2021.687975 (2021).
Brigham and Women’s Hospital and Harvard Medical School Chin Lynda 9 11 Park Peter J. 12 Kucherlapati Raju 13 et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70. https://doi.org/10.1038/nature11412
Marquard, A. M. et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 3, 9. https://doi.org/10.1186/s40364-015-0033-4 (2015).
Funding
This study was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01043754; NRF-2019R1C1C1002830). The funder played no role in study design; data collection, analysis, and interpretation; or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
JK designed the study. TY, BJC, YJC, JL, SGA, and JK contributed to data acquisition. JK and ML analyzed the data. ML, JK, and AL wrote and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, M., Yoo, TK., Chae, B.J. et al. Luminal androgen receptor subtype and tumor-infiltrating lymphocytes groups based on triple-negative breast cancer molecular subclassification. Sci Rep 14, 11278 (2024). https://doi.org/10.1038/s41598-024-61640-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-61640-z
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.