Abstract
Breast cancer is the most common diagnosed cancer, the HER2-positive subtype account for 15% of all breast cancer. HER2-targeted therapy is the mainstay treatment for HER2-positive breast cancer. Cuproptosis is a novel form of programmed cell death, and is caused by mitochondrial lipoylation and destabilization of iron-sulfur proteins triggered by copper, which was considered as a key player in various biological processes. However, the roles of cuproptosis-related genes in HER2-positive breast cancer remain largely unknown. In the present study, we constructed a prognostic prediction model of HER2-positive breast cancer patients using TCGA database. Dysregulated genes for cells resistant to HER2-targeted therapy were analyzed in the GEO dataset. KEGG pathway, GO enrichment and GSEA was performed respectively. The immune landscape of DLAT was analyzed by CIBERSORT algorithm and TIDE algorithm. HER2-positive breast cancer patients with high CRGs risk score showed shorter OS. DLAT was downregulated and correlated with better survival of HER2-positive breast cancer patients (HR = 3.30, p = 0.022). High expressed DLAT was associated with resistant to HER2-targeted therapy. Knocking down DLAT with siRNA increased sensitivity of breast cancer to trastuzumab. KEGG pathway and GO enrichment of DEGs indicated that DLAT participates in various pathways correlated with organelle fission, chromosome segregation, nuclear division, hormone-mediated signaling pathway, regulation of intracellular estrogen receptor signaling pathway, condensed chromosome and PPAR signaling pathway. There was a negative correlation between TIDE and DLAT expression (r = − 0.292, p < 0.001), which means high DLAT expression is an indicator of sensitivity to immunotherapy. In conclusion, our study constructed a four CRGs signature prognostic prediction model and identified DLAT as an independent prognostic factor and associated with resistant to HER2-targeted therapy for HER2-positive breast cancer patients.
Similar content being viewed by others
Introduction
Breast cancer (BC) has become the most frequently diagnosed cancer, and it is also the leading cause of cancer-related death in women worldwide1,2. BC can be divided into four conventional subtypes, Luminal A, Luminal B, human epidermal growth factor 2 (HER2) positive and triple negative breast cancer (TNBC)3. Endocrine therapy is used for individuals with hormone receptor positive disease, chemotherapy is the primary treatment option for TNBC, and HER2-targeted therapy is utilized for patients whose tumors overexpress HER24,5.
About 15% of patients developed HER2-positive BC, which was characterized by a high recurrence rates, aggressive phenotype, and poor survival outcomes6,7. The monoclonal antibody trastuzumab significantly improves the prognosis of HER2-positive individuals8,9,10. Over the past 20 years, significant progress has been achieved in the treatment of HER2-positive BC, and the U.S. Food and Drug Administration (FDA) has approved 7 kinds of HER2-targeted therapies for the treatment of early and/or advanced disease11. These treatment advances have converted into higher cure rates for early-stage disease, as well as significantly longer progression free survival (PFS) and overall survival (OS) in metastatic patients12. Despite these advances, HER2-positive metastatic BC remains an almost invariably fatal disease, and the efficacy of a single HER2-targeted therapy is transient, particularly for patients who relapse after neoadjuvant therapy containing monoclonal antibody, with median PFS of approximately 1 year and less than 1 year in the first and second lines, respectively13. While primary resistance to HER2-targeted therapy is possible, most treatment failures stem from acquired resistance to cell subclones gradually selected over the course of treatment. Primary or secondary resistance to HER2-targeted therapy, however, remains a vexing problem11,14. In fact, BC is a heterogeneous disease with distinct molecular features, biological characteristics and clinical outcomes15,16. In this context, treatments that can induce other forms of regulated cell death (RCD), including apoptosis, ferroptosis, pyroptosis, necroptosis, have emerged as potential strategies to treat drug-resistant BC and improve survival17,18.
Cuproptosis is a recently identified RCD, which is a process occurs via direct binding of copper to lipoylated components of the tricarboxylic acid (TCA) cycle19. The intracellular copper concentration is controlled by copper importers SCL31A1 and copper exporters ATP7A and ATP7B. In addition, serum copper concentrations in BC patients are higher than that in healthy controls and patients with benign breast disease, suggesting that elevated serum copper levels may be associated with an increased risk of BC20. In many other kinds of cancers, like lung cancer21, colorectal cancer22, thyroid cancer23 and prostate cancer24, copper concentrations are also higher than in healthy people. There is increasing evidence suggest that copper as involved in cancer development and spreading (epithelial to mesenchymal transition, angiogenesis), and the responsiveness of many copper complexes to cancer cells has been studied over the past four decades, many of which may become drug options for cancer treatment25. There are 14 cuproptosis-related genes (CRGs) so far. Since the concept of cuproptosis was proposed, CRGs have been considered valuable in predicting prognosis and immune infiltration in patients with BC26,27,28. However, the predictive value may vary among subtypes of BC. Based on CRGs, TNBC patients can be divided into two subtypes, which have significantly different immune invasion characteristics and tumor signaling pathways29. But the studies on the role of cuproptosis in HER2-positive BC lack to some extent.
In our study, we explored the correlation between CRGs with the prognosis of HER2-positive BC through accessing and analyzing the public database, and the prognostic gene set model was constructed. DLAT was identified as a CRGs associated with resistant to HER2-targeted therapy and immunotherapy sensitivity.
Results
Expression determination and survival assessment of cuproptosis-related genes in HER2-positive BC
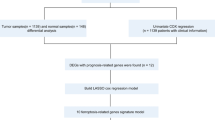
The workflow of the study design was presented in Fig. 1. In this study, we identified 14 genes related to cuproptosis according to the literature30. The expression level of the 14 genes in HER2-positive BC were determined by the TCGA database. As shown in Fig. 2A, nine genes including DLAT, DLD, GLS, LIPT1, MTF1, PDHA1, PDHB, SLC25A3 and SLC31A2 were significantly downregulated, but the other 3 genes including ATOX1, CDKN2A and SLC31A1 were significantly upregulated in HER2-positive BC compared with normal tissues. For FDX1 and LIAS, however, there had no statistical difference between HER2-positive BC and normal breast tissues. Subsequently, the prognostic values of the 14 genes were evaluated in HER2-positive BC patients. The univariate Cox hazard model suggested a higher risk of death in the high expression of DLAT (HR = 3.30, p = 0.022) and SLC25A3 (HR = 2.90, p = 0.039) group (Fig. 2B). The cumulative OS curves provided a visualization of the prognosis with different DLAT and SLC25A3 expression (Fig. 2C).
Expression difference and survival analysis of CRGs in HER2-positive breast cancer. (A) Expression difference of 14 CRGs in HER2-positive breast cancer and normal breast tissues. *p < 0.05; ***p < 0.001; ns, no significance. (B) The forest plot of univariate COX hazard model of the 14 CRGs. *p < 0.05. (C) The OS curves of DLAT and SLC25A3 in patients with HER2-positive breast cancer in the low and high expression group.
The prediction model for HER2-positive BC
Given that CRGs had distinct expression between normal breast tissues and HER2-positive BC, and part of these genes had predictive value for the prognosis of HER2-positive BC patients. We attempted to construct the prognostic gene set for HER2-positive BC using the LASSO method. The prognostic model was established using the 14 cuproptosis related genes in HER2-positive BC patients (Fig. 3A, B). The risk score = (0.1843) * DLAT + (− 0.4056) * SLC31A2 + (0.7899) * SLC25A3 + (0.4987) * ATOX1. The risk score, survival status and expression of patients with HER2-positive BC were shown in Fig. 3C. The further OS curve analysis showed that patients in the low-risk group had a lower risk of death and longer survival time (HR = 0.31, p = 0.026, Fig. 3D).
Construction of the cuproptosis-related prognostic signature. (A) LASSO regression analysis of 14 prognostic CRGs. (B) Cross validation method to select optimal genes. (C) Rank dot, scatter plots and heat map of the model gene expression. (D) Kaplan–Meier analyses of the OS between the high risk and low risk group.
DLAT is the potential gene resistant to HER2-targeted therapy
To identify whether the CRGs have the predictive value for resistance to HER2-targeted therapy, we first analyzed the GEO dataset, GSE12110531. Dysregulated genes for cells resistant to trastuzumab or trastuzumab + pertuzumab in combination were showed in Fig. 4A, B, respectively. Compared with the 14 cuproptosis related genes, only 4 genes showed differently expression in all 3 datasets (Fig. 4C). Further analysis revealed that DLAT was the only gene included both in CRG model genes and resistant genes (Fig. 4D). RT-qPCR found that DLAT expression was significantly down-regulated after transfected with siRNA targeting DLAT (Fig. 4E). MTT assay revealed that DLAT silencing sensitized the SKBR-3 cell to trastuzumab (Fig. 4F).
Selection for the CRGs that related to HER2-targeted therapy. (A) Dysregulated genes for cells resistant to trastuzumab in GSE121105. (B) Dysregulated genes for cells resistant to trastuzumab + pertuzumab in GSE121105. (C) Venn diagram of trastuzumab resistance, trastuzumab + pertuzumab resistance and CRGs. (D) Venn diagram of resistant genes and CRG model genes. (E) The relative expression of DLAT after transfected with si-DLAT. ***p < 0.001. (F) Effect of trastuzumab on growth inhibition in SKBR-3 breast cancer cells transfected with si-DLAT. *p < 0.05; **p < 0.01; ***p < 0.001.
The relationship between DLAT expression and genetic characteristic of BC patients
The genetic characteristic of DLAT was identified. The somatic mutation rate of DLAT was 0.2% (Fig. 5A). The relationship between DLAT and MMR related genes was showed in Fig. 5B, and DLAT has significant correlation with EPCAM, PMS2, MSH6, MSH2 and MLH1. DLAT also had significant correlation with TMB score (r = 0.148, p = 0.049), MSI score (r = − 0.203, p = 0.006) and stemness score (r = 0.299, p < 0.001) (Fig. 5C). By using the ssGESA method, we identified DLAT had positive correlation with DNA replication (r = 0.161, p = 0.032) and tumor proliferation (r = 0.204, p = 0.007), however, DLAT had negative correlation with angiogenesis (r = − 0.197, p = 0.009) (Fig. 5D).
The relationship between DLAT expression and clinical characteristic of BC patients
We analyzed the association between prognosis and DLAT expression in BC patients, however, there had no difference of OS between high or low DLAT expression patients (HR = 1.35, p = 0.08, Fig. 6A). To visualize the attributes of individual patients and display any correlations with survival, we constructed the alluvial diagram (Fig. 6B). The expression level of DLAT in different BC subtype according the PM50 was showed in Fig. 6C, the DLAT had lower expression in Luminal A than Luminal B (p < 0.001), HER2-positive (p < 0.05) and Basal subtype (p < 0.001), while there had no significant difference among Luminal B, HER2-positive and Basal subtype. DLAT had higher expression in T4 than T1&T2&T3 subtype (p < 0.05, Fig. 6D) and higher expression in pre-menopause than post-menopause patients (p < 0.05, Fig. 6E). DLAT also had higher expression in ER negative (p < 0.001) and PR negative patients (p < 0.01), however, there had no difference between HER2-negative and HER2-positive patients (Fig. 6F). In the KM-plotter database, patients in the DLAT low expression group (HR = 1.36, p = 0.031) had significantly longer RFS than patients in the high expression group (Fig. 6G). In the TNM-plotter database, the expression of DLAT had significantly difference between HER2-positive BC and normal breast tissues (p < 0.001, Fig. 6H), and it also varies among normal breast tissues, tumor tissues and metastatic tissues (p < 0.001, Fig. 6I).
The clinical characteristic of DLAT. (A) The OS curves of breast cancer patients with DLAT low and high expression group. (B) The alluvial diagram of DLAT with clinical characteristic. (C) DLAT expression in different PM50 subtype. *p < 0.05; ***p < 0.001; ns, no significance. (D) DLAT expression in different T stage. *p < 0.05. (E) DLAT expression in different menopause status. *p < 0.05. (F) DLAT expression in different ER status, PR status and HER2 status. **p < 0.01; ***p < 0.001; ns No significance. (G) The RFS curves of HER2-positive BC patients with DLAT low and high expression group. (H) DLAT expression between normal breast tissues and HER2-positive BC tissues. (I) DLAT expression among normal breast tissues, HER2-positive BC tissues and metastatic tissues.
GO, KEGG and GESA analysis for DLAT
To further investigate the potential role of DLAT for HER2-positive BC, we divided the HER2-positive BC patients into two groups according to the DLAT expression, namely, DLAT high group and DLAT low group. The upregulated and downregulated genes between the two groups were showed in Fig. 7A, under the condition of given threshold of FDR < 0.05 and fold change > 1. KEGG pathway and GO enrichment of DEGs indicated that DLAT participates in various pathways correlate with organelle fission (GO:0048285), chromosome segregation (GO:0007059), nuclear division (GO:0000280), hormone-mediated signaling pathway (GO:0009755), regulation of intracellular estrogen receptor signaling pathway (GO:0033146), condensed chromosome (GO:0000793) and PPAR signaling pathway (hsa03320) (Fig. 7B, C). By performing GSEA, we further verified the potential roles of DLAT associated molecules in the regulation of cell cycle checkpoints, class I MHC mediated antigen processing presentation, DNA repair, interferon signaling and organelle biogenesis and maintenance (Fig. 7D).
The immune landscape for DLAT
We used the CIBERSORT algorithm to assess the difference of immune infiltration in HER2-positive BC patients with high or low DLAT expression. There is obvious difference in the distribution of 7 out of 22 immunocytes. As compared to the DLAT low group, the infiltration of B cell naïve (p < 0.01) and M2 macrophages (p < 0.001) subtypes was higher, while the infiltrating of B cell memory (p < 0.01), T cell CD8+ (p < 0.05), T cell fh (p < 0.05), Tregs (p < 0.001) and Neutrophil (p < 0.01) subtypes was lower in DLAT high group (Fig. 8A). We analyzed the correlation between DLAT and immune checkpoint inhibitors related genes (Fig. 8B). In addition, the difference in sensitivity to immunotherapy between patients in the DLAT high and low groups was further investigated using the TIDE algorithm. The TIDE algorithm is a recently developed tool for determining the efficacy of tumor immune checkpoint therapy32. In the present study, we found there was a negative correlation between TIDE and DLAT expression (r = − 0.292, p < 0.001, Fig. 8C).
The immune landscape for DLAT. (A) The infiltrating levels of immune cells in high and low DLAT expression groups in HER2-positive breast cancer patients. *p < 0.05; **p < 0.01; ***p < 0.001. (B) The correlation between DLAT and immune checkpoint inhibitors related genes. (C) TIDE predicted response to immunotherapy.
The predicted value of DLAT for HER2-low BC patients
We further established the prognostic nomogram to quantitatively predict the 1-, 2- and 3-year survival probability of HER2-low BC patients (Fig. 9A). The calibration curve of the nomogram demonstrated that it could predict the survival probability relatively well (Fig. 9B). Although there had no difference of OS between DLAT high and low groups for HER2-low BC patients (HR = 2.15, p = 0.24, Fig. 9C). However, the HR+, DLAT low patients had longer OS than HR−, DLAT high patients (HR = 8.61, p = 0.032, Fig. 9D).
The predicted value of DLAT for HER2-low breast cancer patients. (A) The nomogram for predicting HER2-low breast cancer patients’ 1-, 2-, and 3-year OS probability. (B) The 1-, 2-, and 3-year calibration curves of the nomogram. (C) The OS curves of HER2-low breast cancer patients between DLAT low and high groups. (D) The OS curves of HER2-low breast cancer patients between HR+, DLAT low and HR−, DLAT high groups.
Discussion
Cuproptosis is a newly defined RCD, which occurs via direct binding of copper to lipoylated components of the tricarboxylic acid (TCA) cycle and this results in lipoylated protein aggregation and subsequent iron-sulfur cluster protein loss leading to proteotoxic stress and ultimately cell death19. In this study, we identified a novel prognostic model consisting of four CRGs for HER2-positive BC patients. DLAT was downregulated and correlated with better survival of HER2-positive BC patients. High expressed DLAT was associated with resistant to HER2-targeted therapy. However, high expressed DLAT had lower TIDE score, which means it is sensitive to immunotherapy.
The prognostic model for HER2-positive BC was established according to the LASSO method. Four CRGs, named ATOX1, DLAT, SLC31A2 and SLC25A3, were included in this prognostic model. DLAT gene was the E2 core component of the pyruvate dehydrogenase (PDH) mitochondrial enzyme complex, which encoding dihydrolipoamide acetyltransferase. Dihydrolipoamide acetyltransferase accepts acetyl groups and transfers them to coenzyme A33,34. DLAT plays an important role in glucose metabolism and TCA cycle19,35. Goh et al. found that DLAT had higher expression in eleven gastric cancer cells lines compared with non-cancer gastric epithelial cell line HFE145, and pyruvate level was increased when DLAT knockdown with siRNA36. Another research found that DLAT expression have positive relationship with PD-L1 in a clinical hepatocellular carcinoma (HCC) cohort, and DLAT was upregulated and correlated with poor prognosis37. Zhou et al. found that in the HCC cells, the proliferation and invasion ability was decreased once DLAT was knockdown, and the PI3K/Akt or Wnt/β-catenin signaling pathways were also significantly inhibited38. In our study, however, DLAT was downregulated in HER2-positive BC patients compared with normal tissues. Patients in the low DLAT expression group had significantly longer OS than patients in the high expression group. The negative correlation between DLAT expression and infiltrating of B cell memory, CD8+ T cell, fh T cell, Tregs and neutrophil was the unique immune landscape for HER2-positive BC. But a positive correlation between DLAT expression and B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells was found in the HCC cells38. The GSEA verified the potential roles of DLAT associated molecules in the regulation of cell cycle checkpoints, class I MHC mediated antigen processing presentation, DNA repair, interferon signaling and organelle biogenesis and maintenance. DLAT may play distinct roles between HCC and HER2-positive BC.
Several potential mechanisms, such as reduced drug binding to HER2, constitutive activation of HER2 parallel or downstream signaling pathways, metabolic reprogramming or reduced immune system activation, have been identified as primary/secondary resistance to anti-HER2 agents14. Trastuzumab can bind with HER2 extracellular domain IV, which can further prevent the formation of HER2-HER2 homodimers and HER2-HER3 heterodimers39, this also leads to the inhibition of HER2 ectodomain shedding and prevent the expression of the p95-HER2 isoform40. The additional mechanisms including antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Trastuzumab was recognized by Fc receptors of the innate immune system cells, including natural killer cells, antigen presenting cells and effector immune cells, once it bound to HER2 on cancer cell membranes, and this reduced the bounding of trastuzumab with cancer cell41,42. In our study, we found that DLAT high expressed HER2-positive BC patients have shorter OS and associated with resistance to trastuzumab and trastuzumab + pertuzumab. We identified DLAT had positive correlation with DNA replication and tumor proliferation and DLAT also had higher expression in T4 stage than T1&T2&T3 stage. All the evidence suggests that HER2-positive BC patients with high DLAT expression have a higher degree of malignancy and get limited benefits from HER2-targeted therapy. In our vitro experiment, knocking down of DLAT with siRNA significantly improved the treatment effect of trastuzumab. In addition, we found that high expressed DLAT had lower TIDE score, which means it is sensitive to immunotherapy. Immunotherapy has changed the treatment of several cancer types, and it has already been approved as a standard of care for patients with TNBC43,44. Immunotherapy for HER2-positive BC have been an interesting field and it is currently under clinical investigation. Based on the high TMB and high levels of tumor-infiltrating lymphocytes (TILs) of HER2-positive BC, investigating of immunotherapy for HER2-positive BC is feasible45,46. In this study, we found that DLAT also had significant positive correlation with TMB. DLAT high expressed HER2-positive BC patients may benefit from HER2-targeted therapy combined with immunotherapy.
Disruption of mitochondrial activity leading to physiological imbalances and metabolic shifts in the cell, which contribute to cancer progression. One of the most important reasons is mutations in TCA cycle enzymes47,48. However, as part of the TCA cycle enzymes, the somatic mutation rate of DLAT was 0.2% according to this study. The typical PDH deficiency patients were caused by E1α mutations, however, two PDH deficiency patients with DLAT mutation reported by Head et al. were less severely and both have survived well into childhood49. Shatokhina et al. demonstrated that DLAT was down regulated after the control U87 glioma cells were exposed under glutamine deprivation50. In our study, the expression level of DLAT had no difference between HER2-negative and HER2-positive patients. Nevertheless, it was still ambiguous whether DLAT was aberrant regulated in HER2-positive BC patients under some specific conditions. TCA cycle genes including IDH2, SUCLA2, SDHB, SDHD were downregulated after co-expression of HER2 and mucin 1 cytoplasmic domain (MUC1-CD). Interestingly, MUC1-CD induced reduction in TCA cycle genes was found to be significantly associated with poor survivals in patients with HER2-positive BC51. Chang et al. demonstrated that in trastuzumab resistant gastric cancer cell lines (NCI N87R and MKN45R), the activity of GATA binding protein 6 (GATA6) was enhanced prominently. In GATA6 knockout cell lines, the sensitivity to trastuzumab was improved and the mitochondrial function was inhibited52. As DLAT plays an important role in glucose metabolism and TCA cycle, it also acted as a CRGs. In our study, KEGG pathway and GO enrichment of DEGs indicated that DLAT participates in various pathways correlate with organelle fission, chromosome segregation, nuclear division, hormone-mediated signaling pathway, regulation of intracellular estrogen receptor signaling pathway, condensed chromosome and PPAR signaling pathway. Despite the fact that the expression of DLAT had no difference between HER2-negative and HER2-positive BC patients, the exact function of DLAT in HER2-positive BC need further investigation.
In summary, we identified a novel prognostic model consisting of four CRGs for HER2-positive BC patients. In addition, DLAT was further confirmed to be downregulated and correlated with better survival of HER2-positive BC patients. High expressed DLAT related to resistance to HER2-targeted therapy and sensitivity to immunotherapy. However, this study also had some limitations. Firstly, the 14 CRGs were selected from the literature and there may exist some other genes related with cuproptosis, those results we got just applicable at the present stage. Secondly, we did not explore the detailed function of the four CRGs, especially DLAT, in HER2-positive BC. Furthermore, it is unclear how DLAT affects anti-tumor immunity in HER2-positive BC patients.
Methods
Data resource and procession
RNA sequencing data of BC and normal tissues and their corresponding clinical characteristics were obtained from The Cancer Genome Altas (TCGA, https://portal.gdc.cancer.gov/) and the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). All patients included in this study were female. Subsequent analyses were performed with R software (v4.0.3). If not mentioned, p value < 0.05 were considered statistically significant.
Expression analysis and survival analysis of prognostic CRGs in breast cancer
Fourteen genes related to cuproptosis were retrieved from the latest literature30. The differential expression of 14 CRGs between BC and normal tissues was compared with student t-test, including ATOX1, CDKN2A, DLAT, DLD, FDX1, GLS, LIAS, LIPT1, MFT1, PDHA1, PDHB, SLC25A3, SLC31A1, and SLC31A2. Univariate Cox regression analysis was performed to find genes with prognostic value in HER2-positive BC. Survival analyses were performed and Kaplan–Meier survival curves were constructed. The boxplot, forest plot and survival plot were carried out with R packages ‘ggplot2’, ‘forestploter’, and ‘suviminer’.
The establishment of a CRG-based model and prognostic analysis
We established an efficient prediction model using LASSO-Cox analysis according to the methods mentioned in this article53. The risk score was calculated as follows: Riskscore = (0.1843) * DLAT + (− 0.4056) * SLC31A2 + (0.7899) * SLC25A3 + (0.4987) * ATOX1. The coefficient index represents the risk factor, and the gene name represents the expression level of each gene. According to the median risk score, the patients were divided into high risk (risk score > median) and low risk (risk score < median) groups. The LASSO regression, rank dot plot, scatter plots, heat map and survival plot were carried out with R packages ‘ggplot2’, ‘glmnet’, and ‘suviminer’.
Differential expression genes (DEGs) in trastuzumab-resistant datasets
The anti-HER2 therapy datasets were derived from GSE121105 from GEO. We used three groups in this dataset: normal HER2-positive BC cell line (BT474), Trastuzumab resistant cell line (BT474-TR1, BT474-TR2), and both Trastuzumab and Pertuzumab resistant cell line (BT-TPR1, BT-TPR2). Each group contains 3 replicates of sequencing data. The DEGs were selected using the empirical Bayesian approach54. The significance criteria were set as an adjusted p value < 0.001 and an absolute value of Log2 Fold change > 1.5. The volcano plot and Venn diagram were carried out with R packages ‘GEOquery’, ‘limma’, ‘ggplot2’, and ‘VennDiagram’.
The analyses of DLAT’s correlation with genetic characteristics
The genetic characteristics information was obtained from TCGA. The somatic DNA mutations of DLAT were downloaded and visualized using the R packages ‘maftools’. The DNA mismatch repair (MMR) was evaluated with the expression of 5 mismatching repair genes55. The Tumor mutation burden (TMB)56, Microsatellite instability (TSI)57, and cancer stemness score58 were calculated with Spearman’s correlation analysis. The genes related to the DNA replication, tumor proliferation and angiogenesis were picked up from the pathways mentioned in this article59 with R package ‘GSVA’ chosing parameter as method = ‘ssgsea’ and also analyzed by Spearman’s correlation. The scatter plot was carried out with R packages ‘ggstatsplot’.
The analyses of DLAT’s correlation with clinical features
Based on TCGA data, the Kruskal–Wallis test was used to explore the relationships between DLAT and clinical features (ages, pTNM stages, PAM50 subtypes, menopause status, and biomarkers). The boxplot, Sankey diagram, and survival plot were carried out with R packages ‘ggplot2’, ‘ggalluvial’, and ‘suviminer’. The KM-plotter database60 was used to explore the replase free survival (RFS) of HER2-positive BC with different DLAT expression. TNM plotter database61 were used to explore the expression of DLAT in normal breast tissues, HER2-positive BC and metastatic tissues.
The functional enrichment analyses of DLAT
HER2-positive BC samples in the TCGA dataset were divided by DLAT median expression to obtain DEGs. The significance criteria were set as an adjusted p value < 0.001 and an absolute value of Log2 Fold change > 1. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes Genomes (KEGG)62,63 analyses were based on the DEGs. The Gene Set Enrichment Analysis (GSEA) of DLAT was further investigated. All the functional enrichment analyses and visualization were carried out with R packages ‘clusterProfiler’, ‘org.Hs.eg.db’, ‘msigdbr’, ‘GOplot’, and ‘ggplot2’.
The immunological analyses of DLAT in the HER2-positive breast cancer microenvironment
The immune landscape of DLAT was first investigated by calculating by CIBERSORT algorithm64 by using R package ‘immunedeconv’. The immune checkpoint markers were extracted from a previous article65 and analyzed by Spearman’s correlation. The boxplot and heatmap were carried out with R packages ‘ggplot2’and ‘corrplot’. The potential response to immunotherapy was predicted by the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm (http://tide.dfci.harvard.edu/)32. The scatter plot was carried out with R packages ‘ggstatsplot’.
Confirming the prognostic value of DLAT in HER2-low patients
The HER2-low patients were defined as HER2 immunohistochemistry (IHC) results as “1+” in the TCGA dataset. The multivariate Cox regression and corresponding nomogram scoring system were used to develop predictive models of 1-, 3-, and 5-year survival rates. The calibration curve was used to compare the predicted survival rate with the actual one of 1-, 3-, and 5-year survival rates. The nomogram, calibration curve and survival curve were drawn by R package ‘survival’ and ‘rms’.
Cell culture
The human breast cancer cell lines SKBR-3 was obtained from the American Type Culture Collection (ATCC), and cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Sigma-Aldrich, USA) at 37°C in a humidified incubator with 5% CO2.
Cell transfection
To knock down DLAT gene expression, small interfering RNA (siRNAs) were transfected into cell with jetPRIME (Polyplus-transfection Inc, France) according to the Manufacturer’s instructions. Prior to functional studies, we characterized and optimized the knock down of DLAT using 3 different siRNA sequences (si-DLAT-1, si-DLAT-2 and si-DLAT-3) over a concentration of 10, 25, 50 and 100 nM. 50 nM of siRNA was deemed to be optimal for knock down of DLAT. In addition, si-DLAT-1 and si-DLAT-2 were more effective in silencing DLAT expression than si-DLAT-3 (data not shown). The siRNA sequences were purchased from Life Technologies (Thermo Fisher Scientific, Rockford, IL) under the following product codes targeting the respective exons: si-DLAT-1 sequence (HSS102785, targeting exon 1): 5′-UCGCAACAGCGUGACUACAGGGUA-3′; si-DLAT-2 sequence (HSS102786, targeting exon 13): 5′-GGAUAAACUGGUCCCUGCAGAUAAU-3′. Luc control siRNA #5270471 was purchased from Life Technologies, USA.
RNA extraction and RT-qPCR
Total RNA of cells was extracted using Simply P Total RNA Extraction kit (Bioer Technology, China) and subsequently reverse transcribed to complementary deoxyribonucleic acid (cDNA) using PrimerScript™ RT Master Mix (Takara, Shiga, Japan) on a SimpliAmp™ Thermal Cycler (Applied Biosystems, Massachusetts, USA) according to the manufacturer’s instructions. Obtained cDNAs were quantified with a reverse transcription quantitative polymerase chain reaction (RT-qPCR) test labeled with SYBR® green (Invitrogen, California, USA) on LightCycler® 96 (Roche, Mannheim, Germany). The gene-specific primers used were as follows: DLAT forward primer, 5′-CCGCCGCTATTACAGTCTTCC-3′, DLAT reverse primer: 5′-CTCTGCAATTAGGTCACCTTCAT-3′, β-actin forward primer, 5′-CATGTACGTTGCTATCCAGGC-3′; β-actin reverse primer, 5′-CTCCTTAATGTCACGCACGAT-3′. Gene expression levels were normalized to β-actin expression using the 2−ΔCT method. Each cDNA sample was triplicated in 96-microwell plates.
Cell growth assay
Growth inhibition was assessed using the 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation and Cytotoxicity Assay Kit (Beyotime Institute of Biotechnology, Shanghai, China). Cells were diluted in 100 μl/well of maintenance cell culture media and plated in 96-well flat-bottom plates. The number of SKBR-3 cells per well used in the subsequent experiments were 5000. At 24 h after plating, cell culture media were replaced with 10% FBS-containing media with and without trastuzumab, followed by incubation for an additional 120 h. Trastuzumab concentrations ranged from 0.001 to 100 μg/ml. A total of 3 plate wells were set for each experimental point, and all experiments were carried out at least in triplicate. Data are expressed as percentage of growth relative to that of untreated control cells.
Data availability
Any data and R script in this study can be obtained from the corresponding author upon reasonable request. The final manuscript was read and approved by all authors. In this study, publicly available datasets were analyzed. These are available on The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/).
Change history
04 April 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41598-024-58710-7
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Miller, K. D. et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 72, 409–436. https://doi.org/10.3322/caac.21731 (2022).
Goldhirsch, A. et al. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 24, 2206–2223. https://doi.org/10.1093/annonc/mdt303 (2013).
Waks, A. G. & Winer, E. P. Breast cancer treatment: A review. JAMA 321, 288–300. https://doi.org/10.1001/jama.2018.19323 (2019).
Maughan, K. L., Lutterbie, M. A. & Ham, P. S. Treatment of breast cancer. Am. Fam. Phys. 81, 1339–1346 (2010).
Slamon, D. J. et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182. https://doi.org/10.1126/science.3798106 (1987).
Slamon, D. J. et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244, 707–712. https://doi.org/10.1126/science.2470152 (1989).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792. https://doi.org/10.1056/nejm200103153441101 (2001).
Piccart-Gebhart, M. J. et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 353, 1659–1672. https://doi.org/10.1056/NEJMoa052306 (2005).
Romond, E. H. et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 353, 1673–1684. https://doi.org/10.1056/NEJMoa052122 (2005).
Choong, G. M., Cullen, G. D. & O’Sullivan, C. C. Evolving standards of care and new challenges in the management of HER2-positive breast cancer. CA Cancer J. Clin. 70, 355–374. https://doi.org/10.3322/caac.21634 (2020).
Loibl, S. & Gianni, L. HER2-positive breast cancer. Lancet (London, England) 389, 2415–2429. https://doi.org/10.1016/s0140-6736(16)32417-5 (2017).
Cesca, M. G., Vian, L., Cristóvão-Ferreira, S., Pondé, N. & de Azambuja, E. HER2-positive advanced breast cancer treatment in 2020. Cancer Treat. Rev. 88, 102033. https://doi.org/10.1016/j.ctrv.2020.102033 (2020).
Vernieri, C. et al. Resistance mechanisms to anti-HER2 therapies in HER2-positive breast cancer: Current knowledge, new research directions and therapeutic perspectives. Crit. Rev. Oncol. Hematol. 139, 53–66. https://doi.org/10.1016/j.critrevonc.2019.05.001 (2019).
Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 121, 3786–3788. https://doi.org/10.1172/jci60534 (2011).
Testa, U., Castelli, G. & Pelosi, E. Breast cancer: A molecularly heterogenous disease needing subtype-specific treatments. Med. Sci. https://doi.org/10.3390/medsci8010018 (2020).
Saha, T. & Lukong, K. E. Breast cancer stem-like cells in drug resistance: A review of mechanisms and novel therapeutic strategies to overcome drug resistance. Front. Oncol. 12, 856974. https://doi.org/10.3389/fonc.2022.856974 (2022).
Tang, D., Kang, R., Vanden Berghe, T., Vandenabeele, P. & Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 29, 347–364. https://doi.org/10.1038/s41422-019-0164-5 (2019).
Tsvetkov, P. et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375, 1254–1261. https://doi.org/10.1126/science.abf0529 (2022).
Feng, Y. et al. Serum copper and zinc levels in breast cancer: A meta-analysis. J. Trace Elem. Med. Biol. Organ Soc. Miner. Trace Elem. (GMS) 62, 126629. https://doi.org/10.1016/j.jtemb.2020.126629 (2020).
Wang, W. et al. Serum copper level and the copper-to-zinc ratio could be useful in the prediction of lung cancer and its prognosis: A case-control study in Northeast China. Nutr. Cancer 73, 1908–1915. https://doi.org/10.1080/01635581.2020.1817957 (2021).
Sohrabi, M. et al. Trace element and heavy metal levels in colorectal cancer: Comparison between cancerous and non-cancerous tissues. Biol. Trace Elem. Res. 183, 1–8. https://doi.org/10.1007/s12011-017-1099-7 (2018).
Baltaci, A. K., Dundar, T. K., Aksoy, F. & Mogulkoc, R. Changes in the serum levels of trace elements before and after the operation in thyroid cancer patients. Biol. Trace Elem. Res. 175, 57–64. https://doi.org/10.1007/s12011-016-0768-2 (2017).
Saleh, S. A. K., Adly, H. M., Abdelkhaliq, A. A. & Nassir, A. M. Serum levels of selenium, zinc, copper, manganese, and iron in prostate cancer patients. Curr. Urol. 14, 44–49. https://doi.org/10.1159/000499261 (2020).
da Silva, D. A. et al. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 226, 111634. https://doi.org/10.1016/j.jinorgbio.2021.111634 (2022).
Li, J. et al. The cuproptosis-related signature predicts prognosis and indicates immune microenvironment in breast cancer. Front. Genet. 13, 977322. https://doi.org/10.3389/fgene.2022.977322 (2022).
Li, W., Zhang, X., Chen, Y. & Pang, D. Identification of cuproptosis-related patterns and construction of a scoring system for predicting prognosis, tumor microenvironment-infiltration characteristics, and immunotherapy efficacy in breast cancer. Front. Oncol. 12, 966511. https://doi.org/10.3389/fonc.2022.966511 (2022).
Li, Z. et al. Identification of cuproptosis-related subtypes, characterization of tumor microenvironment infiltration, and development of a prognosis model in breast cancer. Front. Immunol. 13, 996836. https://doi.org/10.3389/fimmu.2022.996836 (2022).
Sha, S. et al. Prognostic analysis of cuproptosis-related gene in triple-negative breast cancer. Front. Immunol. 13, 922780. https://doi.org/10.3389/fimmu.2022.922780 (2022).
Ge, E. J. et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat. Rev. Cancer 22, 102–113. https://doi.org/10.1038/s41568-021-00417-2 (2022).
Gale, M. et al. Acquired resistance to HER2-targeted therapies creates vulnerability to ATP synthase inhibition. Cancer Res. 80, 524–535. https://doi.org/10.1158/0008-5472.can-18-3985 (2020).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558. https://doi.org/10.1038/s41591-018-0136-1 (2018).
Patel, M. S. & Roche, T. E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 4, 3224–3233. https://doi.org/10.1096/fasebj.4.14.2227213 (1990).
Kato, M., Chuang, J. L., Tso, S. C., Wynn, R. M. & Chuang, D. T. Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex. EMBO J. 24, 1763–1774. https://doi.org/10.1038/sj.emboj.7600663 (2005).
Solmonson, A. & DeBerardinis, R. J. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 293, 7522–7530. https://doi.org/10.1074/jbc.TM117.000259 (2018).
Goh, W. Q., Ow, G. S., Kuznetsov, V. A., Chong, S. & Lim, Y. P. DLAT subunit of the pyruvate dehydrogenase complex is upregulated in gastric cancer-implications in cancer therapy. Am. J. Transl. Res. 7, 1140–1151 (2015).
Bai, W. D. et al. A novel cuproptosis-related signature identified DLAT as a prognostic biomarker for hepatocellular carcinoma patients. World J. Oncol. 13, 299–310. https://doi.org/10.14740/wjon1529 (2022).
Zhou, Y. et al. Glycolysis-related gene dihydrolipoamide acetyltransferase promotes poor prognosis in hepatocellular carcinoma through the Wnt/β-catenin and PI3K/Akt signaling pathways. Ann. Transl. Med. 10, 1240. https://doi.org/10.21037/atm-22-5272 (2022).
Junttila, T. T. et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell 15, 429–440. https://doi.org/10.1016/j.ccr.2009.03.020 (2009).
Molina, M. A. et al. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 61, 4744–4749 (2001).
Clynes, R. A., Towers, T. L., Presta, L. G. & Ravetch, J. V. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat. Med. 6, 443–446. https://doi.org/10.1038/74704 (2000).
Arnould, L. et al. Trastuzumab-based treatment of HER2-positive breast cancer: An antibody-dependent cellular cytotoxicity mechanism?. Br. J. Cancer 94, 259–267. https://doi.org/10.1038/sj.bjc.6602930 (2006).
Agostinetto, E., Eiger, D., Punie, K. & de Azambuja, E. Emerging therapeutics for patients with triple-negative breast cancer. Curr. Oncol. Rep. 23, 57. https://doi.org/10.1007/s11912-021-01038-6 (2021).
Agostinetto, E. et al. Progress and pitfalls in the use of immunotherapy for patients with triple negative breast cancer. Expert Opin. Investig. Drugs 31, 567–591. https://doi.org/10.1080/13543784.2022.2049232 (2022).
Luen, S., Virassamy, B., Savas, P., Salgado, R. & Loi, S. The genomic landscape of breast cancer and its interaction with host immunity. Breast (Edinburgh, Scotland) 29, 241–250. https://doi.org/10.1016/j.breast.2016.07.015 (2016).
Solinas, C., Carbognin, L., De Silva, P., Criscitiello, C. & Lambertini, M. Tumor-infiltrating lymphocytes in breast cancer according to tumor subtype: Current state of the art. Breast (Edinburgh, Scotland) 35, 142–150. https://doi.org/10.1016/j.breast.2017.07.005 (2017).
Marquez, J. et al. Rescue of TCA cycle dysfunction for cancer therapy. J. Clin. Med. https://doi.org/10.3390/jcm8122161 (2019).
Mullen, A. R. et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388. https://doi.org/10.1038/nature10642 (2011).
Head, R. A. et al. Clinical and genetic spectrum of pyruvate dehydrogenase deficiency: Dihydrolipoamide acetyltransferase (E2) deficiency. Ann. Neurol. 58, 234–241. https://doi.org/10.1002/ana.20550 (2005).
Shatokhina, H. O. et al. ERN1 dependent impact of glutamine and glucose deprivations on the pyruvate dehydrogenase genes expression in glioma cells. Endocr. Regul. 56, 254–264. https://doi.org/10.2478/enr-2022-0027 (2022).
Pang, Z. et al. MUC1 triggers lineage plasticity of Her2 positive mammary tumors. Oncogene 41, 3064–3078. https://doi.org/10.1038/s41388-022-02320-y (2022).
Chang, J., Wang, Q., Bhetuwal, A. & Liu, W. Metabolic pathways underlying GATA6 regulating Trastuzumab resistance in Gastric Cancer cells based on untargeted metabolomics. Int. J. Med. Sci. 17, 3146–3164. https://doi.org/10.7150/ijms.50563 (2020).
Lossos, I. S. et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Engl. J. Med. 350, 1828–1837. https://doi.org/10.1056/NEJMoa032520 (2004).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Li, X., Liu, G. & Wu, W. Recent advances in Lynch syndrome. Exp. Hematol. Oncol. 10, 37. https://doi.org/10.1186/s40164-021-00231-4 (2021).
Thorsson, V. et al. The immune landscape of cancer. Immunity 48, 812-830.e814. https://doi.org/10.1016/j.immuni.2018.03.023 (2018).
Bonneville, R. et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. https://doi.org/10.1200/po.17.00073 (2017).
Lian, H. et al. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol. Oncol. 13, 2227–2245. https://doi.org/10.1002/1878-0261.12557 (2019).
Wei, J. et al. Characterization of glycolysis-associated molecules in the tumor microenvironment revealed by pan-cancer tissues and lung cancer single cell data. Cancers https://doi.org/10.3390/cancers12071788 (2020).
Gyorffy, B. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731. https://doi.org/10.1007/s10549-009-0674-9 (2010).
Bartha, Á. & Győrffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22052622 (2021).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587-d592. https://doi.org/10.1093/nar/gkac963 (2023).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Zeng, D. et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 7, 737–750. https://doi.org/10.1158/2326-6066.cir-18-0436 (2019).
Acknowledgements
We are grateful to our lab members for valuable discussion.
Funding
The conduct of this study was sponsored and funded by National Natural Science Foundation of China (Grant Number 82303865) to Huijuan Dai, the Research Foundation for Talents of Yijishan Hospital of Wannan Medical College (Grant Number YR20220201), Natural Science Research Project of Anhui Educational Committee (Grant Number 2023AH051770) and Foundation for High-level Talent of Provincial Public Medical and Health Institutions (Grant Number GCCRC2022013) to Rui Sha, the Science and Technology Commission of 396 Huaian Municipality, Jiangsu, China (Grant Number HAB202209) to Aijun Sun.
Author information
Authors and Affiliations
Contributions
R.S., X.D. and S.Y. gathered and processed data. R.S. and H.D. drafted the manuscript. L.Y., Z.G. and A.S. had the idea and launched the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the order of the author names, which was incorrectly given as Rui Sha, Xinrui Dong, Shanshan Yan, Huijuan Dai, Liuxia You, Zongjin Guo and Aijun Sun.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sha, R., Dong, X., Yan, S. et al. Cuproptosis-related genes predict prognosis and trastuzumab therapeutic response in HER2-positive breast cancer. Sci Rep 14, 2908 (2024). https://doi.org/10.1038/s41598-024-52638-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-52638-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.