Abstract
The clinical applications of brain age prediction have expanded, particularly in anticipating the onset and prognosis of various neurodegenerative diseases. In the current study, we proposed a deep learning algorithm that leverages brain structural imaging data and enhances prediction accuracy by integrating biological sex information. Our model for brain age prediction, built on deep neural networks, employed a dataset of 3004 healthy subjects aged 18 and above. The T1-weighted images were minimally preprocessed and analyzed using the convolutional neural network (CNN) algorithm. The categorical sex information was then incorporated using the multi-layer perceptron (MLP) algorithm. We trained and validated both a CNN-only algorithm (utilizing only brain structural imaging data), and a combined CNN-MLP algorithm (using both structural brain imaging data and sex information) for age prediction. By integrating sex information with T1-weighted imaging data, our proposed CNN-MLP algorithm outperformed not only the CNN-only algorithm but also established algorithms, such as brainageR, in prediction accuracy. Notably, this hybrid CNN-MLP algorithm effectively distinguished between mild cognitive impairment and Alzheimer’s disease groups by identifying variances in brain age gaps between them, highlighting the algorithm’s potential for clinical application. Overall, these results underscore the enhanced precision of the CNN-MLP algorithm in brain age prediction, achieved through the integration of sex information.
Similar content being viewed by others
Introduction
Chronological aging is intricately linked to several neurodegenerative conditions, including cognitive impairments and dementia1. During the natural aging process, the human brain experiences gray matter (GM) atrophy along with cortical thinning2,3. Despite these commonalities, the aging process of the human brain exhibits significant biological complexity and demonstrates marked inter-individual differences in both its rate and pattern4,5. Furthermore, underlying pathologies may hasten brain aging, and the individual brain aging may be differently influenced by both genetic and environmental factors for each person4,5,6.
Machine learning techniques that utilize brain magnetic resonance imaging (MRI) data can take these variations into account, thereby enhancing the accuracy in predicting an individual's brain age. The estimated brain age at an individual level serves as a personalized indicator for potential brain dysfunction7,8,9. Additionally, the difference between the predicted brain age and the chronological age, referred to as the "brain age gap", has emerged as a promising biomarker for detecting inter-individual differences in brain aging7,8,9. As the positive or negative brain age gap can respectively indicate accelerated or healthy brain aging, individual quantification of this gap may aid in both risk screening and the diagnostic process for neurodegenerative diseases7,9.
As structural MRI of the brain can detect aging-related neuroanatomical changes, such as global GM atrophy, it has been established that the chronological ages of healthy individuals can be accurately estimated using data from structural brain MRI3,5,10. These aging-related changes in brain structures differ according to gender11. Given that individual brain structures reflect both male and female characteristics in complex and dynamic patterns, the quantification of these patterns using machine learning should take into consideration these sex-specific brain characteristics12. Despite the understanding of the sex-specific trajectory of brain aging, there is a paucity of brain age prediction models that incorporate sex information as a relevant feature13. Instead, most of the current algorithms for brain age prediction utilizing structural brain MRI data tend to consider sex information only in the subsequent statistical correction process14.
In order to accurately predict brain age based on structural brain MRI data, the selection of suitable types of input data and algorithms for the optimal learning of normal brain aging patterns is essential9,14. From an algorithmic perspective, high-dimensional regression models employing deep neural networks have increasingly been utilized for brain age estimation7,8,15,16. Additionally, recent deep learning investigations have employed hybrid algorithms, incorporating numerical and/or categorical data to enhance prediction accuracy17,18,19. Specifically, the convolutional neural network (CNN), widely utilized in brain age prediction, has been recognized as optimal for interpreting highly complex brain structures20,21, and the supplementary use of the multi-layer perceptron (MLP) may offer advantages in terms of computing efficiency, depending on the types of input data17,22.
The current study is designed to introduce a novel algorithm that integrates both CNN and MLP algorithms for the prediction of brain age using mixed inputs, including minimally preprocessed T1-weighted images and biological sex information. We hypothesized that this combined CNN-MLP approach may demonstrate superior performance over the CNN-only algorithm, which relies solely on T1-weighted images for predicting brain age. The model's performance was evaluated in the internal validation set (n = 301), and external validation set (n = 645). Additionally, the performance of the combined CNN-MLP algorithm will be juxtaposed with that of the brainageR and pyment algorithms15,23, the latter two being the most widely used and extensively validated algorithms based on structural brain MRI data24,25.
Results
Performance of the brain age prediction algorithms in the training and test sets
The predictive accuracy of both the combined CNN-MLP algorithm and the CNN-only algorithm, tested on the training set (n = 2703) and the test set (n = 301), is detailed in Table 1 and Fig. 1.
Comparison of the performance between the brain age prediction algorithms in internal validation set. Scatter plots show the predicted brain age versus chronological age in the internal validation set (test set) using the combined CNN-MLP algorithm (a, blue dots) and the CNN-only algorithm (b, gray dots). In all plots, the red line indicates a linear regression line and the dashed gray line indicates a y = x line (45-degree line). Abbreviations: CNN, convolutional neural network; MAE, mean absolute error; MLP, multi-layer perceptron; RMSE, root mean squared error; R2, coefficient of determination.
In the training set, a 10-fold cross-validation of the proposed algorithm yielded the following outcomes: The combined CNN-MLP algorithm attained a mean MAE of 3.494 years, a mean RMSE of 4.689 years, and a mean R2 of 0.933. Conversely, the CNN-only algorithm reached a mean MAE of 3.563 years, a mean RMSE of 4.839 years, and a mean R2 of 0.932.
For the test set, the internal validation performance metrics were as follows: the combined CNN-MLP algorithm recorded an MAE of 3.184 years, an RMSE of 4.687 years, and an R2 of 0.936 (Fig. 1a), while the CNN-only algorithm achieved an MAE of 3.342 years, an RMSE of 4.659 years, and an R2 of 0.937 (Fig. 1b).
These results suggest that the combined CNN-MLP method, utilizing both minimally preprocessed T1-weighted images and sex information, outperformed the CNN-only algorithm that used only the minimally preprocessed T1-weighted image.
For further analysis, we assessed the efficiency of a more streamlined model, integrating a linear fully-connected layer at the end of the CNN model, capable of handling sex information. Within the training set, this model’s 10-fold cross-validation produced a mean MAE of 3.674 years, a mean RMSE of 5.042 years, and a mean R2 of 0.926. The test set performance metrics for the CNN integrated with a linear fully-connected layer were as follows: MAE of 3.592 years, RMSE of 4.989 years, and R2 of 0.927.
Performance of the brain age prediction algorithms using the external validation set
The predictive accuracy of the combined CNN-MLP algorithms in the external validation set is detailed in Table 2 and Fig. 2.
Comparison of the performance between the brain age prediction algorithms in external validation set. Scatter plots show the predicted brain age versus chronological age in the external validation set (CamCAN dataset) using the combined CNN-MLP algorithm (a, blue dots), CNN-only algorithm (b, light blue dots), and brainageR algorithm (c, black dots). In both plots, the red line indicates a linear regression line and the dashed gray line indicates a y = x line (45-degree line). Abbreviations: CamCAN, Cambridge Centre for Ageing and Neuroscience; CNN, convolutional neural network; MAE, mean absolute error; MLP, multi-layer perceptron; RMSE, root mean squared error; R2, coefficient of determination.
In the external validation set (n = 645), which was sourced from the Cambridge Centre for Aging and Neuroscience (CamCAN) database (available at http://www.mrc-cbu.cam.ac.uk/datasets/camcan/), the combined CNN-MLP algorithm achieved an MAE of 4.910 years, an RMSE of 6.148 years, and an R2 of 0.891 (Fig. 2a); the CNN-only algorithm achieved an MAE of 5.064 years, an RMSE of 6.295 years, and an R2 of 0.885 (Fig. 2b). These findings consistently demonstrate that the combined CNN-MLP algorithm enhanced the predictive accuracy of brain age in the independent dataset.
We further compared the predictive accuracy of our combined CNN-MLP algorithm with the well-validated brainageR algorithm15 using an external validation set (n = 645) (Table 2, Fig. 2). The performance metrics for the brainageR algorithm included an MAE of 5.360 years, an RMSE of 6.923 years, and an R2 of 0.861 (Fig. 2c). Concurrently, retraining our combined CNN-MLP algorithm with the same inputs as the brainageR model, specifically the normalized gray matter and white matter images, resulted in an MAE of 5.276 years, an RMSE of 6.452 years, and an R2 of 0.879 (Table 2). These outcomes collectively suggest that our CNN-MLP model may offer superior predictive accuracy compared to the brainageR algorithm.
In addition, we compared the CNN-MLP algorithm with the pyment algorithm23, using a newly acquired dataset of healthy subjects (n = 200) from the Alzheimer's Disease Neuroimaging Initiative 1 (ADNI1, available at https://adni.loni.usc.edu/about/adni1/) and Open Access Series of Imaging Studies 1 (OASIS-1, available at http://www.oasis-brains.org/) databases. This selection of a new validation set was necessitated by the prior utilization of the CamCAN dataset, our original external validation set, in the training of the pyment model23. In this comparison, our hybrid CNN-MLP algorithm achieved an MAE of 5.111 years, an RMSE of 6.531 years, and an R2 of 0.919, while the pyment algorithm outperformed the CNN-MLP with an MAE of 4.264 years, an RMSE of 5.664 years, and an R2 of 0.939. The superior performance of the pyment algorithm might be partly attributed to its considerably larger training dataset (n = 53,542), compared to our dataset (n = 2703).
Performance of the brain age prediction algorithms following bias correction
The performance of the combined CNN-MLP algorithm, both before and after the application of bias correction, is detailed in Table 3 and Fig. 3. The application of linear bias correction improved the predictive performance of the combined CNN-MLP algorithm in both internal validation (n = 301) and external validation (n = 645).
Scatter plots of brain age gap versus chronological age without and with linear bias correction. Results of brain age gap (predicted brain age—chronological age) without bias correction (a, c), and with bias correction (b, d). Scatter plots show the brain age gap, predicted by the proposed algorithm, versus chronological age in the test set (a, b), and in the CamCAN dataset (c, d). In all plots, the red lines indicate linear regression lines, and the dashed gray lines indicate ideal estimation references (y = 0). Abbreviations: CamCAN, Cambridge Centre for Ageing and Neuroscience.
In the internal validation, the performance metrics included an MAE of 3.184 years, an RMSE of 4.687 years, and an R2 of 0.936 prior to bias correction (Fig. 3a) and an MAE of 3.134 years, an RMSE of 4.510 years, and an R2 of 0.941 after bias correction (Fig. 3b). In the external validation, the performance metrics were an MAE of 4.910 years, an RMSE of 6.148 years, and an R2 of 0.891 before bias correction (Fig. 3c) and an MAE of 4.313 years, an RMSE of 5.546 years, and an R2 of 0.911 after bias correction (Fig. 3d).
Performance of the brain age prediction algorithms with data augmentation
To evaluate the impact of training set enhancement on brain age prediction accuracy, we conducted supplementary analyses using an augmented dataset. This dataset was randomly augmented with a 30% probability, resulting in the generation of additional synthetic images. The augmentation protocol encompassed 3D rotations within a − 10 to 10° range and translations between − 10 and 10 voxels.
The performance of the algorithms on the internal validation set yielded an MAE of 3.283 years, an RMSE of 4.726 years, and an R2 of 0.932. For external validation, the results showed an MAE of 4.945 years, an RMSE of 6.313 years, and an R2 of 0.885. These findings are detailed in Supplementary Table S3.
Visualization of critical brain regions for age prediction
A global average attention map, obtained from the entire test set (n = 301), revealed pronounced activation in the corpus callosum, internal capsule, and brain regions adjacent to the lateral ventricle (Fig. 4a). These findings suggest that these specific areas contribute more significantly to age prediction compared to other regions of the brain.
Visualization of critical brain regions for age prediction utilizing Grad CAM. (a) A global average attention map was created from the entire test set (n = 301). Regions marked with higher values, closer to red on the map, signify a greater contribution of those regions to age prediction. (b) A gender difference attention map was generated by subtracting the average attention map of female (n = 153) from that of male (n = 148). Regions marked with higher values, closer to red on the map, indicate stronger gender-specific influences of those regions on age prediction. All attention maps are overlaid over the averaged brain MR image from the test set.
The gender difference attention map (Fig. 4b), generated by subtracting the average attention map of females (n = 153) from that of males (n = 148), demonstrated that the regions with the most distinct gender-specific contribution to age prediction were congruent with those highly activated in the global average attention map derived from the total sample.
Application of brain age prediction algorithm to the MCI and AD groups
Employing our combined CNN-MLP algorithm, we estimated the brain age for patients with mild cognitive impairment (MCI, n = 208) and those diagnosed with Alzheimer's disease (AD, n = 172), as depicted in Table 4. The mean (SD) brain age gaps were calculated as 0.413 (3.515) years for the MCI group and 1.393 (3.606) years for the AD group, respectively (Fig. 5). A significant difference in brain age gaps between the MCI and AD groups (t = − 2.673, P = 0.008) was identified. This finding highlights the ability of our current brain age prediction model to efficiently differentiate between the two disease groups, underscoring its clinical relevance.
Distribution of brain age gap by clinical diagnosis. The mean (SD) brain age gaps were 0.413 (3.515) years and 1.393 (3.606) years for the MCI and AD groups, respectively. A significant difference in brain age gaps between the MCI and AD group was identified (t = − 2.673, P = 0.008). Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment; SD, standard deviation.
Discussion
By concatenating sex information with structural brain MRI data, the combined CNN-MLP algorithm exhibited higher accuracy in brain age prediction, in contrast to the CNN-only algorithm that relied solely on T1-weighted images. Furthermore, the combined CNN-MLP algorithm demonstrated superior predictive performance for brain age relative to the previously validated algorithms for brain age prediction, such as the brainageR model15.
In the present study, the hybrid architecture of the CNN-MLP algorithm was effective in achieving high accuracy for brain age prediction, a finding in line with recent research, evidencing enhanced performance and broader applicability through the synergistic use of multiple algorithms to proficiently manage diverse input types17,18,19,26,27,28. Specifically, the concatenating of the CNN algorithm with the MLP algorithm resulted in superior model performance, effectively accommodating factors that influence brain age, such as gender, site identification, and scanner information9,29,30,31. The hybrid CNN-MLP model, adept at merging x-ray images with numerical and categorical medical data, revealed a substantial 5–10% enhancement in discerning COVID-19 infection compared to existing models17. Our supplementary findings highlight the greater efficiency of the hybrid CNN-MLP algorithm over the CNN model augmented with an additional linear fully-connected layer, especially in processing sex information. The MLP algorithm's relatively streamlined structure, in comparison to other deep learning algorithms, may yield benefits such as reduced computational time and load in the creation of combined models22,28. Consequently, in terms of clinical flexibility and scalability, the pairing of CNN and MLP algorithms might offer a strategic advantage in handling complex data, including datasets containing images and varied clinical details17,18,19,26,27,28.
It is noteworthy that the proposed hybrid deep learning model takes into account both sex information and brain structural images when constructing the model. This is in contrast to other brain age prediction models that have subsequently corrected for sex during the validation process14. Given that sex has been shown to affect regional brain volumes11,12,32,33 and neurodegenerative changes34,35, in distinct and influential ways, integrating brain structures and sex information may bolster the model's efficacy in predicting brain age. This idea is supported by the fact that the CNN-MLP algorithm demonstrated superior predictive performance compared to the CNN algorithm, which relied solely on the T1-weighted image22. Our model bears resemblances to the innovative 3D convolutional network, the two-stage-age network (TSAN), which integrates MR images and sex labels as input variables13. However, TSAN diverges from our approach by incorporating a two-stage cascade architecture, wherein the initial age estimate is refined by a secondary network, adding an additional layer of analysis. This refinement enables TSAN to achieve significant accuracy, as evidenced by an MAE of 2.428 using a dataset of 6,586 subjects. To potentially improve our model’s accuracy, we undertook a supplementary analysis by incorporating a two-stage prediction method similar to that of TSAN (Supplementary Fig. S2, Supplementary Table S2). This adaptation of our CNN-MLP algorithm, to include a two-stage prediction process, yielded an MAE of 2.253 years, demonstrating improved performance closely paralleling that of the TSAN model.
Additionally, our findings reveal that the utilization of minimally preprocessed T1-weighted images in the combined CNN-MLP algorithm yielded better results than those of the tissue-segmented T1 images utilized in the brainageR algorithm15. Given the clinical importance of saving time and simplifying neuroimaging preprocessing, the current brain age prediction model, which employs minimally preprocessed T1-weighted images, can be applied efficiently in clinical environments36,37.
Considering that sex information is complexly and variably reflected in regional brain structures11,12,21,32,33, pinpointing the exact brain structural patterns displaying sex effects in influencing model performance has not reached a consensus, and findings have been inconsistent38,39. Within this framework, the present algorithm is able to simultaneously reflect whole-brain structural features to identify the sex-related pattern of aging in the brain, using minimally preprocessed neuroimaging in conjunction with sex information.
The proposed hybrid deep learning model was corrected for linear bias, utilizing individual neuroimaging during the modeling process, which enhanced the predictive accuracy for brain age. Although bias correction is critical for achieving both high accuracy and stability in brain age prediction14,40,41, most statistical corrections have been conducted based on chronological ages following modeling7,40,41,42. In this study, linear bias correction improved the predictive performance, reducing variance; the predicted brain age was refined by subtracting the offset corresponding to the brain age gap40. It may be inferred that linear bias correction can counter underfitting due to regression dilution and the non-Gaussian age distribution of the proposed model. Specifically, an incrementally increased brain age gap at the youngest and oldest extremities, along with a higher prediction error for individuals older than 50 years of chronological age, have been noted due to inter-individual variations in biological aging and biases in linear regression (e.g., linear regression toward the mean, attenuation)43,44.
To identify the brain regions that significantly influence age prediction, we utilized the Grad-CAM, an explainable artificial intelligence method, to create a voxel-wise average attention map45. In line with previous studies46,47,48,49, we discerned that the corpus callosum, internal capsule, and areas near the lateral ventricle were significant contributors to age prediction. Given the established significance of ventricular enlargement and atrophic changes near the lateral ventricle in the brain aging process50,51, these regions likely play a vital role in enhancing model performance.
Moreover, our findings regarding gender differences in the attention maps corroborate previous research on gender-specific aging processes in white matter areas, particularly around the corpus callosum and internal capsule11,32,34. This underscores the value of incorporating sex information into the brain age model to augment its predictive accuracy.
It is important to note that our hybrid CNN-MLP algorithm accurately predicted brain age in healthy individuals and also adeptly differentiated between the two neurodegenerative disease groups, MCI and AD, by identifying variances in their brain age gaps. The extent of brain age gaps for MCI and AD, as determined by our hybrid CNN-MLP model, aligns with that previously documented by Karim et al.52. From a scientific research perspective, using the brain age prediction model to analyze disease groups, especially in computing brain age gaps, greatly enhances our understanding of the model’s clinical implications7,53. Consequently, the current data robustly support the clinical relevance of our hybrid CNN-MLP model, specifically in the field of neurodegenerative diseases.
The following limitations should be considered in interpreting the current results. It is important to understand changes in brain structure and function that are associated with the variations in sex hormones12. Numerous estrogen receptors are found within the central nervous system, hence differences are evident between childbearing-age women and menopausal women54. Specifically, it has been noted that the characteristics of the brain consistently change in tandem with the menstrual cycle55. However, since information such as menopausal status and menstrual cycle of female subjects were not obtained from the database utilized in this study, the related factors potentially impacting prediction performance were not completely accounted for. Therefore, future investigations that include sex hormonal information alongside neuroimaging may offer additional insights into the effects of gender on brain aging54,55.
While the performance of the model that employs the combined CNN-MLP algorithm did exceed that of the CNN-only algorithm, this improvement did not attain statistical significance. Our findings align with numerous previous studies on brain age prediction models, where numerical differences in model performance were noted but without reaching statistical significance, hinting at performance enhancement15,40,42,53. Nonetheless, future research is warranted to confirm the improved performance of the combined CNN-MLP model, incorporating high-resolution structural images and sex information, through more rigorous statistical evaluations15,40,42,53,56,57.
Moreover, recent algorithms that employed more than 10,000 brain images for training have accomplished brain age prediction with an impressive MAE of less than three years23. In line with this, the pyment model, which benefited from a significant training set (n = 53,542), surpassed our CNN-MLP algorithm, which was developed using a considerably smaller training set (n = 2703), in terms of predictive accuracy. Therefore, enriching the training set could potentially boost the performance of our proposed CNN-MLP algorithm in subsequent studies.
While the CamCAN dataset is recognized for its reflection of the general population in terms of demographic variables58,59, it should be noted that the generalizability of the model's performance across varied populations still demands further examination and validation in future investigations.
It is important to underline that our MLP algorithm solely utilized gender information for predicting brain age, not including several vital features such as scanner information or site identification. This limitation was in part due to the absence or ambiguity of relevant information in the available dataset. Considering the proven capability of the MLP algorithm in handling various types of biological information60,61,62, future work should include essential features such as gender, site identification, or scanner information, all known to influence brain age32,33,34,35. The integration of these features into the hybrid CNN-MLP algorithm may notably augment model performance.
It warrants emphasis that future research utilizing the hybrid CNN-MLP algorithm should carefully incorporate both genetic and environmental factors, due to their well-documented impacts on brain aging63,64,65,66. In alignment with this perspective, recent investigations have developed algorithms skilled in processing multimodal data. This approach provides a more comprehensive framework, integrating MRI data with other relevant variables. For example, Qiang et al.64 created an integrated CNN-MLP framework that effectively combined MRI data with clinical and APOE genetic markers, thereby enhancing the diagnostic accuracy for AD. This underscores the potential benefits of augmenting traditional imaging data with genetic and clinical information to enhance model performance. Similarly, Bintsi et al.65 demonstrated improved performance by concurrently integrating imaging and non-imaging variables, such as blood pressure, stroke history, and alcohol consumption, into brain age estimation models. These non-imaging environmental factors have previously been shown to have significant correlations with brain aging65,66. Employing a multimodal approach that considers both imaging and non-imaging genetic/environmental variables has been shown to improve the accuracy of brain age estimation65.
Furthermore, future research involving multimodal neuroimaging (for example, both functional and structural neuroimaging), feature selection, and optimal parameter tuning could refine and optimize the proposed CNN-MLP algorithm67,68,69.
In the current study, the hybrid CNN-MLP algorithm, utilizing the minimally preprocessed T1-weighted images along with sex information, showed higher accuracy in predicting brain age compared to the CNN-only algorithm. These findings may suggest that neuroanatomical changes in brain aging could intertwine with sexually dimorphic clinical features. Accordingly, the proposed CNN-MLP algorithm could broaden our understanding of individual brain aging patterns in the context of both normal and pathological aging and provide critical insights regarding sexually individualized interventions.
Methods
Data collection
The current study included 3004 T1-weighted images of healthy subjects, whose ages ranged from 18.0 to 86.3 years, sourced from various open neuroimaging databases (mean age = 42.1 years, standard deviation [SD] = 18.7; consisting of 1471 men and 1,533 women). We excluded individuals with significant neurological or psychiatric disorders. For the longitudinal databases that contained follow-up brain imaging, only the brain structural MRI images from the baseline assessment were utilized to prevent data leakage between the training and test sets.
The dataset was stratified according to each age bin to ensure an identical age distribution in both the training and test sets. It was randomly divided into the training set (n = 2703) and the test set (n = 301).
The databases included 1000 Functional Connectomes Project (1000 FCP, available at http://fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html)70,71, International Neuroimaging Data-Sharing Initiative (INDI, available at http://fcon_1000.projects.nitrc.org/indi/IndiPro.html)71, Information eXtraction from Images (IXI, available at https://brain-development.org/ixi-dataset), Open Access Series of Imaging Studies 3 (OASIS-3, available at https://oasis-brains.org/)72, OpenNeuro (available at https://openneuro.org/), and Cambridge Centre for Ageing Neuroscience (CamCAN, available at http://www.mrc-cbu.cam.ac.uk/datasets/camcan/)73.
The corresponding Institutional Review Boards of the aforementioned open databases (1000 FCP, INDI, IXI, OASIS-3, OpenNeuro, CamCAN) either provided waivers or granted approval for the submission of anonymized data. Written informed consent was obtained from each subject. This research was conducted in compliance with the Declaration of Helsinki. The databases and detailed information regarding the included subjects are provided in Table 5.
Data preprocessing
Data preprocessing was conducted using Statistical Parametric Mapping (SPM) 12 software (Wellcome Centre for Human Neuroimaging, London, UK). This process involved non-linearly registering T1-weighted images in native space to the Montreal Neurological Institute (MNI) standard space. Such normalization across various scanner types and acquisition protocols ensures consistent model training. The normalization process in SPM12 also incorporated corrections for MR gradient field deviations, employing "bias regularization" and "bias FWHM" options74,75. Subsequently, the processed images were resampled to a voxel resolution of 1.5 mm using cubic spline interpolation, yielding a field-of-view of 105 × 127 × 105.
Brain age prediction algorithms
In this study, we employed a three-dimensional (3D) CNN architecture, utilizing minimally preprocessed T1-weighted images with a dimension of 105 × 127 × 105 for brain age estimation15. This architecture consists of sequential convolutional blocks, each encompassing a 3D convolution layer, batch normalization layer, rectified linear unit (ReLU) activation function, and a max pooling layer with a stride of two. The initial block incorporated eight feature channels, while subsequent blocks double this number to better capture the intricate nuances of brain structures 15.
Following the convolutional blocks, the output from the final block was flattened and directed into a dense layer with sixty-four neurons and ReLU activation. This was then succeeded by a batch normalization layer, a dropout layer with a rate of 0.3, and another dense layer with sixteen neurons, again activated by ReLU.
The MLP architecture, formulated to process categorical sex information, integrated a dense layer with sixteen neurons activated by ReLU, followed by another dense layer with four neurons, also under ReLU activation.
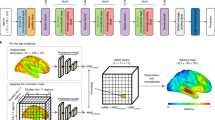
To create the combined CNN-MLP algorithm, the outputs from the concatenation layer were used as inputs. This concatenated input underwent processing through a dense layer with four neurons activated by ReLU, followed by an additional dense layer with a single neuron. Lastly, a linear activation function was applied to this final dense layer, deriving the predicted brain age. A schematic representation of the proposed architecture is depicted in Fig. 6.
Overview of the proposed combined CNN-MLP algorithm for brain age prediction. The CNN architecture, designed for minimally preprocessed T1-weighted images, consists of repeated convolutional blocks, each with 3D convolutional layers, batch normalization, ReLU activations, and max pooling. After these blocks, the sequence includes a flattening layer, two dense layers interspersed with ReLU activations, batch normalization, and a dropout layer. The MLP, tailored for categorical sex information, features dense layers with ReLU activations. Both algorithms' outputs are merged by a concatenation layer, processed through two dense layers, with the final layer using a linear activation for brain age prediction. Abbreviations: 3D, 3-dimensional; BatchNorm, batch normalization; CNN, convolutional neural network; Conv, Convolution; MaxPool, max pooling; MLP, multi-layer perceptron; ReLU, rectified linear unit.
The proposed algorithm was refined through hyperparameter tuning, a method renowned for boosting the accuracy of the brain age prediction model by adjusting key hyperparameters like batch size, epoch, learning rate, and neural network structural variables76,77. Hyperparameter tuning involves the utilization of different optimizers to stabilize the pattern of model updates76. Specifically, in this study, five optimizers were implemented across two combinations of learning rates and decay values: a learning rate of 0.01 with a decay of 0.003, and a learning rate of 0.001 with a decay of 0.0003. Among the investigated optimizers—adaptive gradient (Adagrad)78, adaptive moment estimation (Adam)79, Nesterov accelerated gradient (NAG)80, root mean square propagation (RMSprop)80, and stochastic gradient descent (SGD)81—the 10-fold cross-validation using Adam, with a learning rate of 0.001 and a decay of 0.0003, yielded the most favorable results. Comprehensive results for each of the five optimizers are detailed in Supplementary Table S1. It should be noted that, due to GPU constraints, the model was trained with a batch size of 16.
In addition, we constructed a CNN-only algorithm, trained exclusively with the minimally preprocessed T1-weighted images by using Adam with a learning rate of 0.001 and a decay of 0.0003, for the purpose of comparing its performance with the proposed combined CNN-MLP algorithm.
Training and testing
To evaluate the performance of each algorithm, we utilized mean absolute error (MAE), root mean squared error (RMSE), and the coefficient of determination (R2) as performance metrics.
In this study, a 10-fold cross-validation scheme was applied to compare the performances of different methods: each algorithm was trained on nine randomly selected subsets, and then validated on the final subset, referred to as the validation set. The optimal algorithm was identified by evaluating the average performance metrics in 10-fold cross-validation.
Utilizing a computational framework comprising two NVIDIA Titan Xp GPUs with 12 GB memory, the training time for the CNN-MLP algorithm was approximately 6.94 h, whereas the CNN-only algorithm necessitated 5.28 h for training.
External validation
The external validation of the proposed algorithms was performed using an independent dataset from the CamCAN (available at http://www.mrc-cbu.cam.ac.uk/datasets/camcan/)73. Recognized for its approximate reflection of the broader UK demographic profile, this dataset is deemed less biased and more generalizable58,59. Due to these attributes, the CamCAN set has been the preferred choice for external validation in numerous previous studies regarding brain age prediction models58,59,82,83,84. Specifically, the dataset, consisting of 645 individuals, demonstrated a balanced distribution of age (mean age = 54.7 years, SD = 18.6 years, range = 18.5–88.9 years, Supplementary Fig. S1) and gender (319 men, 49.5%, mean age = 55.1 years, SD = 18.4 years; 326 women, 50.5%, mean age = 54.3 years, SD = 18.8 years), enhancing its suitability for this study. We further validated our proposed model, the combined CNN-MLP algorithm, by contrasting its performance with well-established brain age prediction algorithm packages, specifically brainageR15. We selected brainageR for performance comparison because of the comparable size of its training dataset (brainageR, n = 3377 vs. our study, n = 2703) and its proven high, well-validated performance, making it a suitable benchmark85. According to Cole et al.15, the brainageR model was constructed using a computational setup that incorporated four NVIDIA Titan X GPUs. While their study15 did not specify the exact training duration, the application of Gaussian process regression (GPR) is known to reduce computational time compared to certain other deep learning algorithms with a similar level of performance.
In addition, we compared our model performance with another model, the pyment model23. The training process of the pyment model spanned approximately 70 h when using two NVIDIA V100 GPUs with 32 GB memory23. We selected it primarily for comparison because of its utilization of the CNN algorithm, a feature aligned with our current study. However, it is important to note that the pyment model was developed using a significantly larger, multisite dataset (n = 53,542), and thus surpassed various brain aging models, including ours, with an MAE of 2.4723,30,86. For this comparison, we employed a new, independent dataset comprising 200 healthy individuals (mean age = 57.6 years, SD = 23.0 years, range = 18.0–90.0 years; consisting of 93 men and 107 women) sourced from the ADNI1 and OASIS-1 databases, as the CamCAN dataset had been previously used in training the pyment model.
Bias correction
The phenomenon of underfitting is frequently observed in brain age prediction models and can be attributed to factors such as regression dilution and non-Gaussian age distribution. Therefore, in the current study, a linear bias correction method40 predicted on the chronological age was employed to diminish the variance and enhance the prediction performance. The procedure entailed the following steps: Initially, the relationship between the offset, derived from the brain age gap (defined as the difference between the predicted brain age and the corresponding chronological age), and chronological age was established. Subsequently, the predicted brain age was refined by subtracting the identified offset.
Visualization of critical brain regions for age prediction
To explore the specific brain regions that notably contribute to brain age prediction, we incorporated the explainable AI technique, gradient-weighted class activation mapping (Grad-CAM), into the CNN algorithm45,87. This approach facilitates the visualization of essential brain regions that are integral to the model's performance, employing a heat map44. Although Grad-GAM was originally devised for classification tasks45, we adapted it for regression algorithms, consistent with previous literature88,
In this particular application, we utilized Grad-GAM within a three-dimensional space to generate attention maps for individual brain images of the test set (n = 301), all registered to the MNI standard template. The values within attention maps were normalized within a range of 0–1, with a higher value denoting a more considerable contribution of a specific region to the overall brain age prediction. A global average voxel-wise attention map was subsequently created by averaging the individual attention maps.
To examine the influence of gender information on predictive performance, we crafted global average voxel-wise attention maps for both male (n = 148) and female (n = 153) samples in the test set. These attention maps could reveal gender-specific vital brain regions for age prediction. In our study, we visualized the gender-specific contributions of brain regions to age prediction by computing the differences between male and female attention maps. These difference values were normalized within a range of 0–1 and illustrated as a voxel-wise map. A higher value suggests a more marked gender difference in the contribution of brain regions to age prediction, possibly reflecting an augmented influence of gender information on age prediction.
Brain age estimation in patients with MCI and AD
To investigate the clinical applicability of our brain age prediction model further, we employed the combined CNN-MLP algorithm to estimate the brain age in patients diagnosed with MCI (n = 208, MCI group) and AD (n = 172, AD group). The data utilized for these analyses were sourced from the ADNI1 database (available at https://adni.loni.usc.edu/about/adni1/)89. We determined the brain age gap, defined as the difference between chronological age and estimated brain age, for both the MCI and AD groups. Subsequently, we compared these brain age gaps between the two groups using an independent t-test.
Data availability
The datasets analyzed during the study are available in the following sources: 1000 Functional Connectomes Project (1000 FCP, available at http://fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html); International Neuroimaging Data-sharing Initiative (INDI) Prospective Data Sharing Samples (available at http://fcon_1000.projects.nitrc.org/indi/IndiPro.html); Information eXtraction from Images (IXI, available at https://brain-development.org/ixi-dataset/); Open Access Series of Imaging Studies (OASIS, available at http://www.oasis-brains.org/); Cambridge Centre for Aging and Neuroscience (CamCAN, available at https://www.cam-can.org/); Alzheimer’s Disease Neuroimaging Initiative 1 (ADNI 1, available at https://adni.loni.usc.edu/).
References
Hou, Y. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 (2019).
Scahill, R. I. et al. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch. Neurol. 60, 989–994 (2003).
Storsve, A. B. et al. Differential longitudinal changes in cortical thickness, surface area and volume across the adult life span: Regions of accelerating and decelerating change. J. Neurosci. 34, 8488–8498 (2014).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Rose, N. The human sciences in a biological age. Theory Cult. Soc. 30, 3–34 (2013).
Chen, B. H. et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging 8, 1844 (2016).
Cole, J. H. & Franke, K. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci. 40, 681–690 (2017).
Franke, K., Luders, E., May, A., Wilke, M. & Gaser, C. Brain maturation: Predicting individual BrainAGE in children and adolescents using structural MRI. NeuroImage 63, 1305–1312 (2012).
Franke, K., Ziegler, G., Klöppel, S., Gaser, C., Alzheimer’s Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. NeuroImage 50, 883–892 (2010).
Sowell, E. R., Thompson, P. M. & Toga, A. W. Mapping changes in the human cortex throughout the span of life. Neuroscientist 10, 372–392 (2004).
Coffey, C. E. et al. Sex differences in brain aging: A quantitative magnetic resonance imaging study. Arch. Neurol. 55, 169–179 (1998).
Cosgrove, K. P., Mazure, C. M. & Staley, J. K. Evolving knowledge of sex differences in brain structure, function, and chemistry. Biol. Psychiatry 62, 847–855 (2007).
Cheng, J. et al. Brain age estimation from MRI using cascade networks with ranking loss. IEEE Trans. Med. Imaging 40, 3400–3412 (2021).
Niu, X., Zhang, F., Kounios, J. & Liang, H. Improved prediction of brain age using multimodal neuroimaging data. Hum. Brain Mapp. 41, 1626–1643 (2020).
Cole, J. H. et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 163, 115–124 (2017).
Dinsdale, N. K. et al. Learning patterns of the ageing brain in MRI using deep convolutional networks. NeuroImage 224, 117401 (2021).
Ahsan, M. M., Alam, E., Trafalis, T. & Huebner, P. Deep MLP-CNN model using mixed-data to distinguish between COVID-19 and Non-COVID-19 patients. Symmetry 12, 1526 (2020).
Desai, M. & Shah, M. An anatomization on breast cancer detection and diagnosis employing multi-layer perceptron neural network (MLP) and Convolutional neural network (CNN). Clin. eHealth 4, 1–11 (2021).
Zhang, C. et al. A hybrid MLP-CNN classifier for very fine resolution remotely sensed image classification. ISPRS J. Photogramm. Remote Sens. 140, 133–144 (2018).
Bae, J. B. et al. Identification of Alzheimer’s disease using a convolutional neural network model based on T1-weighted magnetic resonance imaging. Sci. Rep. 10, 22252 (2020).
Besson, P., Parrish, T., Katsaggelos, A. K. & Bandt, S. K. Geometric deep learning on brain shape predicts sex and age. Comput. Med. Imaging Graph. 91, 101939 (2021).
Karlik, B. & Olgac, A. V. Performance analysis of various activation functions in generalized MLP architectures of neural networks. Int. J. Artif. Intell. 1, 111–122 (2011).
Leonardsen, E. H. et al. Deep neural networks learn general and clinically relevant representations of the ageing brain. NeuroImage 256, 119210 (2022).
Bacas, E. et al. Probing multiple algorithms to calculate brain age: Examining reliability, relations with demographics, and predictive power. Hum. Brain Mapp. 44, 3481–3492 (2023).
Clausen, A. N. et al. Assessment of brain age in posttraumatic stress disorder: Findings from the ENIGMA PTSD and brain age working groups. Brain Behav. 12, e2413 (2022).
Gavrishchaka, V., Yang, Z., Miao, R. & Senyukova, O. Advantages of hybrid deep learning frameworks in applications with limited data. Int. J. Mach. Learn. Comput. 8, 549–558 (2018).
Kuo, C.-Y. et al. Improving individual brain age prediction using an ensemble deep learning framework. Front. Psychiatry 12, 626677 (2021).
Zhang, S. & Niu, Y. LcmUNet: a lightweight network combining CNN and MLP for real-time medical image segmentation. Bioeng. 10, 712 (2023).
Jónsson, B. A. et al. Brain age prediction using deep learning uncovers associated sequence variants. Nat. Commun. 10, 5409 (2019).
Holm, M. C. et al. Linking brain maturation and puberty during early adolescence using longitudinal brain age prediction in the ABCD cohort. Dev. Cogn. Neurosci. 60, 101220 (2023).
Bashyam, V. M. et al. MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain 143, 2312–2324 (2020).
Xu, J. et al. Gender effects on age-related changes in brain structure. Am. J. Neuroradiol. 21, 112–118 (2000).
Becker, J. B. et al. Strategies and methods for research on sex differences in brain and behavior. Endocrinology 146, 1650–1673 (2005).
Fjell, A. M. et al. Minute effects of sex on the aging brain: A multisample magnetic resonance imaging study of healthy aging and Alzheimer’s disease. J. Neurosci. 29, 8774–8783 (2009).
Vegeto, E. et al. The role of sex and sex hormones in neurodegenerative diseases. Endocr. Rev. 41, 273–319 (2020).
Singh, S. P. et al. 3D deep learning on medical images: A review. Sensors 20, 5097 (2020).
Wood, D. A. et al. Accurate brain-age models for routine clinical MRI examinations. NeuroImage 249, 118871 (2022).
Anderson, N. E. et al. Machine learning of brain gray matter differentiates sex in a large forensic sample. Hum. Brain Mapp. 40, 1496–1506 (2019).
Eliot, L., Ahmed, A., Khan, H. & Patel, J. Dump the “dimorphism”: Comprehensive synthesis of human brain studies reveals few male-female differences beyond size. Neurosci. Biobehav. Rev. 125, 667–697 (2021).
Beheshti, I., Nugent, S., Potvin, O. & Duchesne, S. Bias-adjustment in neuroimaging-based brain age frameworks: A robust scheme. NeuroImage Clin. 24, 102063 (2019).
Treder, M. S. et al. Correlation constraints for regression models: Controlling bias in brain age prediction. Front. Psychiatry 12, 615754 (2021).
Le, T. T. et al. A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Front. Aging Neurosci. 10, 317 (2018).
Hutcheon, J. A., Chiolero, A. & Hanley, J. A. Random measurement error and regression dilution bias. Bmj 340, c2289 (2010).
Nesselroade, J. R., Stigler, S. M. & Baltes, P. B. Regression toward the mean and the study of change. Psychol. Bull. 88, 622 (1980).
Selvaraju, R. R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision 618–626 (2017).
Bermudez, C. et al. Anatomical context improves deep learning on the brain age estimation task. Magn. Reson. Imaging 62, 70–77 (2019).
Bintsi, K. M., Baltatzis, V., Hammers, A. & Rueckert, D. Interpretability of Machine Intelligence in Medical Image Computing, and Topological Data Analysis and Its Applications for Medical Data 65–74 (Springer, 2021).
Hepp, T. et al. Uncertainty estimation and explainability in deep learning-based age estimation of the human brain: Results from the German National Cohort MRI study. Comput. Med. Imaging Graph. 92, 101967 (2021).
Levakov, G., Rosenthal, G., Shelef, I., Raviv, T. R. & Avidan, G. From a deep learning model back to the brain—identifying regional predictors and their relation to aging. Hum. Brain Mapp. 41, 3235–3252 (2020).
Resnick, S. M., Pham, D. L., Kraut, M. A., Zonderman, A. B. & Davatzikos, C. Longitudinal magnetic resonance imaging studies of older adults: a shrinking brain. J. Neurosci. 23, 3295–3301 (2003).
Mu, Q., Xie, J., Wen, Z., Weng, Y. & Shuyun, Z. A quantitative MR study of the hippocampal formation, the amygdala, and the temporal horn of the lateral ventricle in healthy subjects 40 to 90 years of age. Am. J. Neuroradiol. 20, 207–211 (1999).
Karim, H. T. et al. Independent replication of advanced brain age in mild cognitive impairment and dementia: detection of future cognitive dysfunction. Mol. Psychiatry 27, 5235–5243 (2022).
Cole, J. Steps towards clinical application of the brain age paradigm. Biol. Psychiatry 91, S3–S4 (2022).
Hara, Y., Waters, E. M., McEwen, B. S. & Morrison, J. H. Estrogen effects on cognitive and synaptic health over the lifecourse. Physiol. Rev. 95, 785–807 (2015).
Pletzer, B., Harris, T. & Hidalgo-Lopez, E. Subcortical structural changes along the menstrual cycle: Beyond the hippocampus. Sci. Rep. 8, 16042 (2018).
Baecker, L. et al. Brain age prediction: A comparison between machine learning models using region-and voxel-based morphometric data. Hum. Brain Mapp. 42, 2332–2346 (2021).
Lones, M. A. How to avoid machine learning pitfalls: A guide for academic researchers. arXiv:2108.02497. https://doi.org/10.48550/arXiv.2108.02497 (2021).
Henson, R. N. et al. Multiple determinants of lifespan memory differences. Sci. Rep. 6, 32527 (2016).
Liu, X., Tyler, L. K., Davis, S. W., Rowe, J. B. & Tsvetanov, K. A. Cognition’s dependence on functional network integrity with age is conditional on structural network integrity. Neurobiol. Aging 129, 195–208 (2023).
Mahmud, M., Kaiser, M. S., Hussain, A. & Vassanelli, S. Applications of deep learning and reinforcement learning to biological data. IEEE Trans. Neural. Netw. Learn. 29, 2063–2079 (2018).
Deepika, D. & Balaji, N. Effective heart disease prediction using novel MLP-EBMDA approach. Biomed. Signal Process. Control 72, 103318 (2022).
So, A., Hooshyar, D., Park, K. W. & Lim, H. S. Early diagnosis of dementia from clinical data by machine learning techniques. Appl. Sci. 7, 651 (2017).
Burgos, N. et al. Deep learning for brain disorders: From data processing to disease treatment. Brief. Bioinform. 22, 1560–1576 (2021).
Qiang, Y. R. et al. Diagnosis of Alzheimer’s disease by joining dual attention CNN and MLP based on structural MRIs, clinical and genetic data. Artif. Intell. Med. 145, 102678 (2023).
Bintsi, K. M. et al. Syntax of referencing. Multimodal brain age estimation using interpretable adaptive population-graph learning. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2023 195–204 (Springer, 2023).
Cole, J. H. Multimodality neuroimaging brain-age in UK biobank: Relationship to biomedical, lifestyle, and cognitive factors. Neurobiol. Aging 92, 34–42 (2020).
Bergstra, J., Bardenet, R., Bengio, Y. & Kégl, B. Algorithms for hyper-parameter optimization. Adv. Neural. Inf. Process. Syst. 24, 1–9 (2011).
Liem, F. et al. Predicting brain-age from multimodal imaging data captures cognitive impairment. NeuroImage 148, 179–188 (2017).
Song, F., Guo, Z. & Mei, D. Feature selection using principal component analysis. In International Conference on System Science, Engineering Design and Manufacturing Informatization Vol. 11656344 27–30 (2010).
Biswal, B. B. et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U. S. A. 107, 4734–4739 (2010).
Mennes, M., Biswal, B. B., Castellanos, F. X. & Milham, M. P. Making data sharing work: The FCP/INDI experience. NeuroImage 82, 683–691 (2013).
LaMontagne, P. J. et al. OASIS-3: Longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. MedRxiv https://doi.org/10.1101/2019.12.13.19014902 (2019).
Taylor, J. R. et al. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) data repository: Structural and functional MRI, MEG, and cognitive data from a cross-sectional adult lifespan sample. NeuroImage 144, 262–269 (2017).
Ashburner, J. et al. SPM12 Manual (Wellcome Centre for Human Neuroimaging, Institute of Neurology, UCL, 2014).
Ganzetti, M., Wenderoth, N. & Mantini, D. Quantitative evaluation of intensity inhomogeneity correction methods for structural MR brain images. Neuroinformatics 14, 5–21 (2016).
Yu, T. & Zhu, H. Hyper-parameter optimization: A review of algorithms and applications. arXiv:2003.05689. https://doi.org/10.48550/arXiv.2003.05689 (2020).
Smith, S. L., Kindermans, P. J., Ying, C. & Le, Q. V. Don’t decay the learning rate, increase the batch size. arXiv:1711.00489. https://doi.org/10.48550/arXiv.1711.00489 (2017).
Lydia, A. & Francis, S. Adagrad—an optimizer for stochastic gradient descent. Int. J. Inf. Comput. Sci. 6, 566–568 (2019).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv:1412.6980. https://doi.org/10.48550/arXiv.1412.6980 (2014).
Ruder, S. An overview of gradient descent optimization algorithms. arXiv:1609.04747. https://doi.org/10.48550/arXiv.1609.04747 (2016).
Loshchilov, I. & Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. arXiv:1608.03983. https://doi.org/10.48550/arXiv.1608.03983 (2016).
Lancaster, J., Lorenz, R., Leech, R. & Cole, J. H. Bayesian optimization for neuroimaging pre-processing in brain age classification and prediction. Front. Aging Neurosci. 10, 28 (2018).
Newcombe, V. F. et al. Post-acute blood biomarkers and disease progression in traumatic brain injury. Brain 145, 2064–2076 (2022).
Feng, X. et al. Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiol. Aging 91, 15–25 (2020).
de Lange, A. M. G. et al. Mind the gap: Performance metric evaluation in brain-age prediction. Hum. Brain Mapp. 43, 3113–3129 (2022).
Khayretdinova, M. et al. Predicting age from resting-state scalp EEG signals with deep convolutional neural networks on TD-brain dataset. Front. Aging Neurosci. 14, 1367 (2022).
Cortez, P. & Embrechts, M. J. Using sensitivity analysis and visualization techniques to open black box data mining models. Inf. Sci. 225, 1–17 (2013).
Wang, J. et al. Gray matter age prediction as a biomarker for risk of dementia. Proc. Natl. Acad. Sci. U. S. A. 116, 21213–21218 (2019).
Aisen, P. S. et al. Clinical core of the Alzheimer’s Disease Neuroimaging Initiative: Progress and plans. Alzheimers Dement. 6, 239–246 (2010).
Acknowledgements
We thank the open databases used in our study: the 1000 Functional Connectomes Project; International Neuroimaging Data-sharing Initiative Prospective Data Sharing Samples; Information eXtraction from; Open Access Series of Imaging Studies; Cambridge Centre for Aging and Neuroscience; Alzheimer's Disease Neuroimaging Initiative 1.
Funding
This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2020M3E5D9080555 and 2020R1A2C2005901), grant funded by the Ministry of Education (2020R1A6A1A03043528) and Creative & Basic Technology Research Project funded by the Electronics and Telecommunications Research Institute (Grant No. 23YS1110).
Author information
Authors and Affiliations
Contributions
S.Y. and J.H. conceptualized the study; S.Y. and I.K.L. acquired funding; Y.J., E.N., S.Y., and H.J. participated in original drafting, data visualization, and formal analysis; I.K., J.K., and S.O. participated in investigation and data curation. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Joo, Y., Namgung, E., Jeong, H. et al. Brain age prediction using combined deep convolutional neural network and multi-layer perceptron algorithms. Sci Rep 13, 22388 (2023). https://doi.org/10.1038/s41598-023-49514-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-49514-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.