Abstract
Alterations in mTOR signalling molecules, including RICTOR amplification, have been previously described in many cancers, particularly associated with poor prognosis. In this study, RICTOR copy number variation (CNV) results of diagnostic next-generation sequencing (NGS) were analysed in 420 various human malignant tissues. RICTOR amplification was tested by Droplet Digital PCR (ddPCR) and validated using the “gold standard” fluorescence in situ hybridisation (FISH). Additionally, the consequences of Rictor protein expression were also studied by immunohistochemistry. RICTOR amplification was presumed in 37 cases with CNV ≥ 3 by NGS, among these, 16 cases (16/420; 3.8%) could be validated by FISH, however, ddPCR confirmed only 11 RICTOR-amplified cases with lower sensitivity. Based on these, neither NGS nor ddPCR could replace traditional FISH in proof of RICTOR amplification. However, NGS could be beneficial to highlight potential RICTOR-amplified cases. The obtained results of the 14 different tumour types with FISH-validated RICTOR amplification demonstrate the importance of RICTOR amplification in a broad spectrum of tumours. The newly described RICTOR-amplified entities could initiate further collaborative studies with larger cohorts to analyse the prevalence of RICTOR amplification in rare diseases. Finally, our and further work could help to improve and expand future therapeutic opportunities for mTOR-targeted therapies.
Similar content being viewed by others
Introduction
The mammalian target of rapamycin (mTOR) has multifunctional effects on several cellular processes of tumours (e.g. proliferation, growth, motility, protein synthesis, transcription and autophagy) by integrating signals from complex cellular networks, including metabolic adaptation mechanisms. Alterations in the mTOR signalling genes are common in malignancies (e.g. high frequency of PIK3CA, PTEN and TSC1/2 mutations)1. The altered activity of two complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), has been described in many cancers, especially in patients with unfavourable prognoses2,3.
mTORC2 plays an essential role in cell differentiation, survival, growth, migration and maintenance of the actin cytoskeleton, mainly through phosphorylation of Akt (at Ser473), SGK1 and PKCα4. Rictor (rapamycin-insensitive companion of mTOR) is a characteristic actin co-ordinating scaffold protein of the mTORC2—instead of Raptor in the mTORC1—and its primary function is to assemble the complex. Regarding the function of mTOR complexes, it has been hypothesised that the RICTOR amplification could contribute to cancer progression and metastasis, through mTORC2-mediated functions in the regulation of cancer-related biochemical pathways (e.g. Wnt/β-catenin and MAPK/ERK pathways)5.
RICTOR amplification results in an increased expression of the Rictor protein and is usually associated with an increased mTORC2 complex activity. Moreover, RICTOR amplification or overexpression has significant clinical importance and is associated with poor prognosis and shorter overall survival in certain neoplasms6. Based on mutation databases, RICTOR amplification, a known mTOR pathway-activating oncogenic alteration, occurs in ~ 5% of certain solid tumours (e.g. lung, melanoma, endometrial, colorectal and gastric cancers). The significance of RICTOR amplification or Rictor protein overexpression has been described in the lung7,8, breast9,10, skin11 and gastrointestinal cancers12,13,14,15,16. Additionally, increased mTORC2 activity, could offer alternative therapeutic advantages and allow for personalised therapy as a general consequence of RICTOR amplification and their associated signalling alterations.
Over the last few decades, clinical phase trials examining the administration of phosphoinositide 3-kinase (PI3K)/mTOR pathway inhibitors have demonstrated little significant success; therefore, only a limited number of PI3K/mTOR inhibitors have been introduced into clinical practice. Even though rapamycin, an eponymous inhibitory molecule of mTOR kinase, has been isolated from microbial-derived antibiotics and was first introduced in 197517. Rapamycin (or sirolimus) and rapalogs (e.g. everolimus, temsirolimus) inhibit mTORC1 activity. Many studies have shown that high mTORC2 activity and/or Rictor overexpression may correlate with therapy (including rapalog) resistance. Certain mTORC1/2 (both mTORC1 and mTORC2) and dual (both mTOR and other kinase) inhibitors have already been developed and involved in clinical phase II trials (such as vistusertib and sapanisertib). These inhibitors have shown promising results in lung cancer and other solid tumours18,19. Most of these inhibitors are currently used in personalised and/or combined therapies (with conventional chemotherapy/radiotherapy); the development of these inhibitors and their administration are still ongoing20.
Targeted therapies need reliable molecular markers to predict sensitivity. RICTOR gene amplification, other rare RICTOR mutations, epigenetic and/or signalling alterations may initiate over-representation of the mTORC2 complex linked to mTORC2 hyperactivity. These could be predictive markers for the PI3K/mTOR/Akt signalling inhibitor therapy. The above-summarised alterations can be tested using various methods: a. fluorescence in situ hybridisation (FISH) as a “gold standard” diagnostic method to identify RICTOR amplification8,21,22; b. identification of RICTOR sequence and copy number variations (CNVs) using sequencing (e.g. next-generation sequencing—NGS) and Droplet Digital PCR (ddPCR); c. analyses of Rictor expression or mTORC2 activity by Rictor and Phospho-Ser473-Akt (an activated mTORC2 target protein) immunohistochemistry (IHC). A combination of these established technologies can provide a complete analytical platform to determine genetic and protein expression alterations of RICTOR and their signalling consequences.
Our study aimed to validate potential RICTOR amplification detected by NGS in different malignant tumours using FISH, ddPCR; and compare the detected RICTOR amplification with in situ Rictor and Phospho-Ser473-Akt protein expressions obtained by IHC.
Methods
Case selection
Between 2018 and 2022, formalin-fixed and paraffin-embedded (FFPE) tumour tissue samples from 420 patients (mainly digestive system tumours, central nervous system tumours, soft tissue and bone tumours; see Table 1, Supplementary Table 1 for details) were sequenced using diagnostic NGS at the Department of Pathology and Experimental Cancer Research, Semmelweis University (Budapest, Hungary).
Next-generation sequencing
Diagnostic NGS was performed using previously described methods23. Before DNA isolation tumour cell percentages (TC%) of studied samples were estimated by pathologists (samples with TC% < 20% were excluded). Genomic DNA was isolated using the QIAmp DNA FFPE Tissue Kit (QIAGEN GmbH, Hilden, Germany). Illumina TruSight Oncology 500 High Throughput assay (TSO500) library preparation workflow and NGS on Illumina NextSeq 2000 platform with 101 cycles of paired-end sequencing was performed according to the manufacturer’s protocol.
Bioinformatic analysis was performed using Illumina TruSight Oncology 500 Local App v2.1. FASTQ generation was performed by BCL-convert (Binary Base Call) software after raw BCL files were downloaded. Using the Burrows-Wheeler Aligner and SAMtools utility, the sequence alignment to the hg19 reference genome was performed. Duplicate reads were removed by read collapsing analysis, marked by unique molecular identifiers. CNV calling was performed by CRAFT. Cases with normalised RICTOR CNV ≥ 3 were selected (the normalised CNV is 2 for cells without gene amplification and somatic cells containing the “normal”, non-amplified 2 gene copy) for our examinations.
RICTOR FISH and ddPCR analyses
FISH was performed using 4-μm thick FFPE tissue sections. After deparaffinisation and citrate pre-treatment, samples were digested (37 °C, 15 min, 10% pepsin). Hybridisation was performed at 85 °C, 10 min and then at 37 °C overnight using ZytoLight SPEC RICTOR/5q31.1 Dual Color Probe (ZytoVision GmbH; Bremerhaven, Germany). Washing steps (0.4 × SSC/0.1% NP-40 at 72 °C, 5 min and 2 × SSC/0.1% NP-40 at RT, 5 min) and DAPI (Vector Laboratories, Inc. Newark, CA, USA) counterstaining were carried out.
Representative areas were selected and evaluated by counting 30–30 nuclei in at least two regions of the tumour using the Nikon Eclipse E600 fluorescence microscope. The mean of the signal numbers per nucleus and RICTOR/5q31.1 ratio were determined, where RICTOR/5q31.1 ratio ≥ 2 was considered positive for RICTOR amplification (the RICTOR/5q31.1 ratio is 1 in cells without gene amplification).
DNA previously isolated for NGS analysis was used for RICTOR gene copy number analysis by ddPCR. The PCR reaction was conducted from 50 ng extracted DNA with a RICTOR FAM probe (Unique Assay ID: dHsaCNS608884235, Bio-Rad Laboratories, Inc.; Hercules, CA, USA) and an AP3B1 HEX probe (Unique Assay ID: dHsaCP2500348, Bio-Rad Laboratories, Inc.) which were described previously21. Droplets were generated using Bio-Rad Automated Droplet Generator, and the emulsified PCR reactions were run on Bio-Rad C1000 Touch Thermal Cycler (95 °C for 10 min, 40 cycles of 94 °C for 30 s, 60 °C for 1 min, and 98 °C for 10 min). Droplets were read and analysed using Bio-Rad QX200 Droplet Reader and QuantaSoft Software (version 1.7).
Data were normalised to the percentage of TC% of FFPE samples using the formula24:
Normalised RICTOR/AP3B1 ratio ≥ 2 was defined as RICTOR amplification (the RICTOR/AP3B1 ratio is 1 for cells without gene amplification and somatic cells containing the “normal”, non-amplified 2 gene copy). This formula was not used below 40% tumour purity to exclude false results.
Immunohistochemistry
IHC was performed on 4-μm thick FFPE tissue sections. After deparaffinisation, endogenous peroxidase blocking and antigen retrieval (10 mM citrate buffer, pH 6), slides were incubated with primary antibodies: anti-Rictor (Bethyl Laboratories, Inc.; Montgomery, TX, USA; A500-002A; 1:500), anti-Rictor (Cell Signaling; Danvers, MA, USA; #2140; 1:200), and anti-Phospho-Ser473-Akt (Cell Signaling, #4060; 1:100). Immunohistochemical reactions were visualised by Novolink Polymer (Leica Biosystems; Deer Park, IL, USA), followed by Dako Liquid DAB + Substrate Chromogen System (Agilent Technologies, Inc.; Santa Clara, CA, USA) and haematoxylin counterstaining.
Two independent investigators evaluated the IHC results on digitalised slides using CaseViewer software (version: 2.3, 3DHistech; Budapest, Hungary). The H-score method was used for evaluation as described previously25. H-scores were calculated by multiplying the percentage of positive cells by staining intensity (0, 1+, 2+, 3+). H-score < 100 was considered low, and H-score ≥ 100 was regarded as a high expression for both Rictor and Phospho-Ser473-Akt.
Analysis of RICTOR alteration frequency in genomic databases
The data were collected from The Cancer Genome Atlas (TCGA, PanCancer Atlas Studies) and MSK MetTropism, downloaded via cBioPortal (www.cbioportal.org, accessed on 05 Jan 2023)26,27. Furthermore, CNVIntegrate, the first database that stores CNVs from cancer patients and provides statistical comparisons between copy number frequencies in different ethnic populations (www.cnvintegrate.cgm.ntu.edu.tw, accessed on 05 Jan 2023) was also used28.
Statistical analysis
Statistical analyses were carried out using IBM SPSS Statistics (version 25; SPSS Inc.; Armonk, NY, USA). Spearman’s rank correlation was used to assess the correlation between the RICTOR/5q31.1 ratio detected by FISH, the RICTOR/AP3B1 ratio detected by ddPCR, and Rictor (both antibodies) and Phospho-Ser473-Akt expression detected by immunohistochemistry (H-scores). A p-value < 0.05 was considered statistically significant.
Ethics approval and patient consent to participate statement
The archived tissue samples were used with the approval of the Hungarian Scientific Council National Ethics Committee for Scientific Research (No. 7/2006) and the Institutional Ethical Review Board (SE KREB-216/2020).
Patient consent was waived due to the retrospective nature of molecular analyses performed.
Results
Selecting potential RICTOR-amplified cases using TSO500 results
Based on TSO500 results, cases with normalised RICTOR CNV ≥ 3 were selected for further analyses. Potential RICTOR amplification was presumed in 37 (37/420) cases. The median (range) of normalised RICTOR CNV of these selected cases was 3.87 (3.00–7.03). Distribution of 37 cases regarding their origin: digestive system (11/37), female genital (7/37), central nervous system (6/37), soft tissue and bone (4/37), thoracic (4/37), head and neck (2/37), breast tumours (2/37) and tumours of endocrine organs (1/37). Gender and age characteristics of these patients: 54% (20/37) female and 46% (17/37) male; 65% (24/37) younger than 65 years (detailed information in Table 2).
In 37 selected cases, FISH and ddPCR were used to test RICTOR amplification, while IHC was applied for evaluating Rictor and Phospho-Ser473-Akt overexpression.
FISH analysis was also applied to assess RICTOR amplification. The median (range) of RICTOR/5q31.1 was 1.76 (0.94–3.29). RICTOR amplification (defined as RICTOR/5q31.1 ≥ 2) were validated in 16 cases (16/37). Moreover, the median (range) of the normalised RICTOR/AP3B1 ratio was 1.79 (0.44–5.58), and RICTOR amplification (defined as RICTOR/AP3B1 ≥ 2) was observed in 11 cases (11/37) using ddPCR.
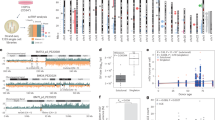
Expressions of Rictor and Phospho-Ser473-Akt were analysed by IHC. Two Rictor antibodies from different companies (Bethyl and Cell Signaling) were applied. High expression of Rictor was detected in 14 (14/37) and 12 (12/37) cases with Bethyl and Cell Signaling antibodies, respectively. High Phospho-Ser473-Akt expression was detected in 7 cases (7/37). Representative images of RICTOR-amplified/non-amplified and high/low protein expressions of IHC stainings are shown in Fig. 1.
Representative images of RICTOR-amplified/non-amplified cases by FISH (a) and the high/low expressions of IHC stainings (b) in the studied tumour samples. The presented squamous cell carcinoma of the lung (case #36) showed RICTOR amplification with high Rictor and high Phospho-Ser473-Akt protein expressions. The other tubo-ovarian high-grade serous carcinoma (case #23) represents no amplification without increased Rictor and Phospho-Ser473-Akt expressions.
In FISH analysis green and orange signals indicate the RICTOR gene and the control locus region (5q31.1); and the scale bars indicate 5 μm/ 50 μm for FISH and IHC, respectively.
Analysis of NGS-predicted RICTOR CNVs, FISH, ddPCR results and in situ expression of characteristic mTORC2 protein (Rictor) and mTORC2 activity marker (Phospho-Ser473-Akt)
Results of RICTOR amplification and/or protein expression analyses were compared to RICTOR CNV numbers obtained by NGS. It was not surprising that: a. NGS-predicted CNV ≥ 5 has the best predictive value for RICTOR amplification among other CNVs (4/5); b. we found relatively more cases with positive RICTOR amplification by FISH among samples with NGS-predicted CNV ≥ 4 (9/12). Surprisingly, lower NGS CNV group (3.00–3.99; 25/37) cases could also be validated by FISH (7/25) or were found amplified by ddPCR (4/25). Additionally, ddPCR confirmed only 11 amplified cases among these 16 validated ones; 4 cases were associated with the lower NGS CNV group (Table 3).
IHC results were also analysed and compared with FISH-validated RICTOR amplification status. The combined evaluation of all three immunostainings associated well with the validated RICTOR amplification. 13 validated RICTOR-amplified cases (13/16) showed high expression with Rictor and/or Phospho-Ser473-Akt immunostainings (at least with one antibody staining) (Table 4).
The combined evaluation of the three different antibody stainings showed high specificity in highlighting RICTOR amplification. The positivity with at least one antibody staining (Rictor and/or Phospho-Ser473-Akt) compared to FISH amplification results showed 81% (13/16) specificity and 66% (14/21) sensitivity in detecting protein expression alterations associated with RICTOR amplification in the studied 37 cases.
Finally, we also assessed the correlations between FISH, ddPCR ratios and immunohistochemistry score results. A strong positive correlation was observed between the RICTOR/5q31.1 ratio detected by FISH and the RICTOR/AP3B1 ratio detected by ddPCR (p = 0.004, Spearman’s R = 0.491). Indeed, Rictor H-scores using antibodies from different distributors (Bethyl and Cell Signaling) also showed a strong positive correlation (p = 0.000, Spearman’s R = 0.639). However, Phospho-Ser473-Akt H-scores correlated only with Rictor H-scores detected by the Cell Signaling antibody (p = 0.002, Spearman’s R = 0.528). No significant correlation was observed between single IHC stainings and FISH or ddPCR results.
Our NGS results and publicly available RICTOR alteration frequency data
We compared our NGS CNV results with publicly available datasets. We used two databases for comparative evaluation: CNVIntegrate and cBioPortal with two datasets (MSK MetTropism and TCGA PanCancer Atlas). A similar frequency of RICTOR gene alterations was found in several tumour types (Table 5). Our data and the MSK MetTropism dataset found the best correlation regarding tumour types (e.g. breast, hepatobiliary tract, female genital, and head and neck tumours). Among our studied cases, the highest percentage of potential RICTOR-amplified cases was found in central nervous system tumours (1.43%), and the lowest was in tumours of the hepatobiliary tract, stomach, endometrium and adrenal gland (0.24%).
It is important to note that the three databases evaluated RICTOR alteration frequency differently. CNVIntegrate (unknown case number; data from the COSMIC dataset) considers an NGS CNV ≥ 2 as an amplified case. The TCGA PanCancer Atlas (n = 10,967) also rates putative copy number alterations ≥ 2 and the MSK MetTropsim (n = 25,775) above 2.29. In our study, cases (n = 37) with CNV ≥ 3 were considered to be set for evaluation.
Genes with altered copy numbers that co-occur with RICTOR CNV
In the validated RICTOR amplification cases, several additional amplifications were detected by FISH for diagnostic purposes. These cases carried some further amplifications. The co-detected amplifications were as follows: 1 case with ERBB2 amplification, 1 case with EGFR amplification, 1 case with ALK amplification, 1 case with ERBB2 amplification and CDK4 amplification, and 1 case with co-occurring ERBB2, MET, ALK and CDK4 amplification.
The NGS results also highlighted some potentially co-occurring altered genes (with CNV ≥ 3) in the validated RICTOR amplification cases. These are as follows: AKT2 (3/16), BRCA1 (1/16), CCND1 (5/16), CCNE1 (2/16), CDK4 (3/16), CDK6 (2/16), EGFR (3/16), ERBB2 (3/16), ERBB3 (1/16), ERCC1 (1/16), ERCC2 (1/16), ESR1 (1/16), FGF3 (3/16), FGF4 (1/16), FGF10 (2/16), FGF19 (2/16), FGFR1 (3/16), FGFR3 (2/16), FGFR4 (3/16), JAK2 (1/16), KIT (2/16), KRAS (2/16), MDM2 (1/16), MDM4 (4/16), MET (3/16), MYC (8/16), MYCL (1/16), NMYC (1/16), PDGFRA (2/16), PIK3CA (2/16), PIK3CB (1/16), RET (2/16), RPS6KB1 (2/16), TFRC (1/16). Among these, FGFR4 and FGF10 are also located on chromosome 5, and the distance between RICTOR (5p13.1) and FGF10 (5p12) genes is relatively low.
Previously undescribed tumour types with RICTOR amplification
We could validate RICTOR amplification in 6 main tumour types with 14 diagnoses (Table 6). There were several tumour diagnoses (n = 10) where our FISH analyses were the first to validate RICTOR amplification. Based on mTOR activity alterations (Rictor overexpression or increased RICTOR CNV, without validated amplification), many of these tumour types have been identified by others as potential mTOR inhibitor targets. These are the following entities: Her2− invasive breast carcinoma of no special type, astrocytoma, glioblastoma, gallbladder adenocarcinoma, pancreatic neuroendocrine tumour, endometrial carcinosarcoma, undifferentiated endometrial carcinoma, oral squamous cell carcinoma, dedifferentiated liposarcoma, squamous cell carcinoma of the lung. Additionally, mTOR/Rictor expression and genetic alterations (e.g. amplification) have not been previously studied and described in pancreatic neuroendocrine tumours and endometrial carcinosarcomas, these are newly validated RICTOR-amplified entities in our study. These cases showed RICTOR amplification by FISH and ddPCR; case #19 with pancreatic neuroendocrine tumour (RICTOR/5q31.1 ratio was 2.47 and normalised RICTOR/AP3B1 ratio was 4.83) and case #20 with endometrial carcinosarcoma (RICTOR/5q31.1 ratio was 2 and normalised RICTOR/AP3B1 ratio was 3.21), as well.
Discussion
The role of molecular diagnostics has recently increased significantly in personalised, molecular-targeted therapy. Sequencing of genomic regions (or even the entire genome) can reveal genomic alterations, confirm base substitutions, deletions and insertions; and highlight potential copy number changes, and gene rearrangements. Comprehensive genetic profiling using NGS requires special attention in pathology practice and laboratory work. These molecular diagnostic results are associated with many challenging difficulties. The validation and interpretation of NGS results are time-consuming and require additional costs, which are critical for clinical treatment decisions51. Besides the well-known targetable genetic alterations, it would be helpful to highlight certain tumour types where some additional targets can be used regarding available NGS results and their validated targets.
In our work, NGS revealed RICTOR CNVs were further analysed in various tumour samples, and RICTOR amplified cases were validated by the “gold standard” FISH, and studied by other molecular and in situ analyses. Potential RICTOR amplification was presumed in 37 cases by NGS and validated in 16 cases by FISH. ddPCR was less sensitive, confirming only 11 RICTOR-amplified cases. Based on these results, the specificity and sensitivity of NGS and ddPCR could not allow for replacing the FISH but could help highlight potential RICTOR-amplified cases. The 14 different cancer diagnoses described with validated RICTOR amplification underline its importance in a wide range of tumours, including some rare malignancies. The already published research works were reviewed to collect data (and referred to in Table 6) where RICTOR amplification or overexpression was detected and described in different tumour types. Regarding these, our newly validated RICTOR amplification results highlighted the following: a. there are several associations between our results and the available data about RICTOR alterations; b. we could validate RICTOR amplification first by FISH in several main tumour types, where previously potentially targetable Rictor overexpression or increased NGS CNV had been highlighted; c. among the 16 FISH-validated cases, pancreatic neuroendocrine tumour and endometrial carcinosarcoma were described for the first time in our study as new, previously not studied RICTOR-amplified entities (neither the RICTOR gene alteration nor the Rictor expression changes have been previously studied in these tumour types).; d. interestingly, RICTOR amplification was not validated in 4 presumed tubo-ovarian high-grade serous carcinoma cases, e. RICTOR amplification was validated in all enrolled lung adenocarcinomas (n = 3).
Our screened patient population had an unusual distribution regarding the Hungarian centralisation, an above-average prevalence of rare malignancies, central nervous system tumours (CNS), as well as soft tissue and bone tumours and paediatric cases (all paediatric tumour samples are sent to Semmelweis University). It should be noted that only a few lung cancers (e.g. lung adenocarcinoma, squamous cell carcinoma of the lung) were included in our recent study (most lung adenocarcinoma cases are tested on Illumina TruSight Tumor 170 panel). We have previously contributed to RICTOR amplification analysis of small cell lung carcinoma cases, and the Hungarian lung adenocarcinoma cohort has already been characterised for mTORC1/2 activity by our research8,49. Among the paediatric RICTOR CNV-selected patients (5/37), 2 CNS tumour cases were validated with RICTOR amplification. This could be particularly interesting in childhood glioblastoma and astrocytoma regarding described mTOR and mTORC2 hyperactivity in CNS malignancies52,53. Phase trials showed low effectiveness of PI3K/Akt/mTOR inhibitors54. However, neither mTORC2 complex hyperactivity nor RICTOR amplification has been studied before treatment decisions.
Regarding the newly validated RICTOR-amplified cases, the potential role of high mTORC2 activity has already been described in glioblastoma, astrocytoma and invasive breast carcinoma of no special type. Targeting PI3K/AKT/mTOR signalling pathway mutations with inhibitors could benefit patients diagnosed with these entities10,55,56,57. Additionally, mTOR inhibitors and specific targeting of Rictor modifications have been very effective in experimental glioblastoma models58,59,60. In ~ 50–60% of gallbladder adenocarcinomas, mTOR hyperactivity has also been described in association with poor prognosis (but RICTOR alterations were not analysed)61,62,63. The revealed undifferentiated endometrial carcinoma case in our cohort underlines the importance of previously described mTORC2 activity as a risk/prognostic factor43 and a phase I study with sapanisertib showing anti-tumour activity in certain renal and endometrial cancer cases64. RICTOR-amplified cases of dedifferentiated liposarcoma, oral squamous cell carcinoma and lung squamous cell carcinoma confirm the importance of rapalogs and dual inhibitors as treatment options in these malignancies65,66,67. Additionally, PIK3CA and/or EGFR amplification and Rictor overexpression in head and neck squamous cell carcinoma and lung squamous cell carcinoma, and the RICTOR amplification detected in our study are associated with the fact that mTOR inhibitors are under investigation in these tumours45,68.
There has been some success with mTOR inhibitors in treating pancreatic neuroendocrine tumours and heavily pre-treated endometrial carcinosarcomas in clinical trials. Still, no data is specific to RICTOR amplification in these malignancies19,69,70,71,72. Additionally, mTOR inhibitors have been tested in combination with gemcitabine73, and in association with PIK3CA mutations, samotolisib showed some efficacy and a manageable safety profile in endometrial carcinosarcomas74. RICTOR gene and expression alterations have not been previously reported in pancreatic neuroendocrine tumours and endometrial carcinosarcomas. Therefore, our is the first study about high mTORC2 activity with validated RICTOR amplification in these entities.
Combined with the validated RICTOR amplification, increased expression of Rictor and Phospho-Ser473-Akt were confirmed as targetable signalling alterations. There were only 3 cases (3/16) where FISH-validated RICTOR amplification was not associated with altered Rictor and/or Phospho-Ser473-Akt protein levels. Possible causes of these discrepancies could be explained by epigenetic modifications and complex biological mechanisms (e.g. post-translational regulatory mechanisms)4,75,76,77,78,79,80,81,82,83; and further technical issues (e.g. biopsy sample handling, antibody specificity and sensitivity differences). These results suggest that the applied immunostainings could be helpful in pre-screening cases before RICTOR FISH analysis. Both RICTOR FISH and the above-mentioned IHCs would be beneficial in better patient selection before targeting mTOR/mTORC1 hyperactivity. Corresponding to our findings, some studies have suggested that RICTOR amplification and/or Rictor overexpression might be predictive markers to identify cancer patients who will respond to dual mTORC1/2 inhibitors18,84. Besides the allosteric mTOR inhibitors (sirolimus, everolimus and temsirolimus), numerous PI3K/Akt/mTOR inhibitors have been under development. mTOR kinase inhibitors (e.g. sapanisertib, vistusertib); dual PI3K, mTOR inhibitors (e.g. paxalisib, samotolisib) and Akt inhibitors (e.g. ipatasertib, capivasertib) are all in active phase 2 and 3 trials85. Nevertheless, the recent monotherapies have low overall survival benefits. The selection of optimal mTOR inhibitor combinations with chemo/radiotherapy in selected patient populations needs to be considered, however, increased toxicity of combination therapy may limit the success of these treatments86.
A biomarker-driven umbrella study which selected patients based on the presence of RICTOR amplification demonstrates a promising strategy for personalised treatments and identifies patients who are most likely to respond to targeted therapy (vistusertib)87. These underline the significance of validated RICTOR amplification targets in small cell lung cancers or other malignancies.
In conclusion, there is no consensus on the actual predictive value of the NGS-predicted CNV. Our results could highlight RICTOR amplification in a wide range of heavily pre-treated and/or difficult-to-treat malignancies. Our findings also demonstrate the importance of RICTOR amplification validation, especially in cases where tumours may have been inappropriately studied using conventional techniques. Additionally, our RICTOR amplification validated cases with 3 ≤ CNV < 4 characteristics could highlight that lower CNV scores should be also considered in further analyses. Finally, RICTOR FISH and IHC stainings (Rictor, Phospho-Ser473-Akt) are highly reliable and cost-effective validation methods before the administration of targeted therapy using mTORC1/2 inhibitors in various cancers.
Data availability
Publicly available datasets were analysed in this study; The Cancer Genome Atlas (TCGA, PanCancer Atlas Studies) and MSK MetTropism, downloaded via cBioPortal (www.cbioportal.org, accessed on 05 Jan 2023). Furthermore, we also used CNVIntegrate (www.cnvintegrate.cgm.ntu.edu.tw, accessed on 05 Jan 2023). Additionally, the original contributions presented in the study are available in the article/Supplementary Material provided. For further inquiries, please contact the corresponding author.
References
Tian, T., Li, X. & Zhang, J. mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int. J. Mol. Sci. 20, 755 (2019).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Sebestyén, A. et al. The role of metabolic ecosystem in cancer progression—Metabolic plasticity and mTOR hyperactivity in tumor tissues. Cancer Metast. Rev. 40, 989–1033 (2021).
Fu, W. & Hall, M. N. Regulation of mTORC2 signaling. Genes 11, 1045 (2020).
Shaw, R. J. & Cantley, L. C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441, 424–430 (2006).
Gkountakos, A. et al. Unmasking the impact of Rictor in cancer: Novel insights of mTORC2 complex. Carcinogenesis 39, 971–980 (2018).
Krencz, I., Sebestyen, A. & Khoor, A. mTOR in lung neoplasms. Pathol. Oncol. Res. 26, 35–48 (2020).
Krencz, I. et al. Correlation between immunohistochemistry and RICTOR fluorescence in situ hybridization amplification in small cell lung carcinoma. Hum. Pathol. 93, 74–80 (2019).
Wazir, U., Newbold, R. F., Jiang, W. G., Sharma, A. K. & Mokbel, K. Prognostic and therapeutic implications of mTORC1 and Rictor expression in human breast cancer. Oncol. Rep. 29, 1969–1974 (2013).
Morrison Joly, M. et al. Rictor/mTORC2 drives progression and therapeutic resistance of HER2-amplified breast cancers. Cancer Res. 76, 4752–4764 (2016).
Jebali, A., Battistella, M., Lebbé, C. & Dumaz, N. RICTOR affects melanoma tumorigenesis and its resistance to targeted therapy. Biomedicines 9, 1498 (2021).
Jiang, W. J. et al. RICTOR expression in esophageal squamous cell carcinoma and its clinical significance. Med. Oncol. 34, 32 (2017).
Driscoll, D. R. et al. mTORC2 signaling drives the development and progression of pancreatic cancer. Cancer Res. 76, 6911–6923 (2016).
Joechle, K. et al. Role of mammalian target of rapamycin complex 2 in primary and secondary liver cancer. World J. Gastrointest. Oncol. 13, 1632–1647 (2021).
Wang, F. et al. Overexpression of Rictor protein and Rictor-H. pylori interaction has impact on tumor progression and prognosis in patients with gastric cancer. Folia Histochem. Cytobiol. 58, 96–107 (2020).
Bian, Y. et al. Elevated Rictor expression is associated with tumor progression and poor prognosis in patients with gastric cancer. Biochem. Biophys. Res. Commun. 464, 534–540 (2015).
Sehgal, S. N., Baker, H. & Vézina, C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot. (Tokyo) 28, 727–732 (1975).
Cheng, H. et al. RICTOR Amplification defines a novel subset of patients with lung cancer who may benefit from treatment with mTORC1/2 inhibitors. Cancer. Discov. 5, 1262–1270 (2015).
Rajdev, L. et al. A phase II study of sapanisertib (TAK-228) a mTORC1/2 inhibitor in rapalog-resistant advanced pancreatic neuroendocrine tumors (PNET): ECOG-ACRIN EA2161. Investig. New Drugs 40, 1306–1314 (2022).
Conciatori, F. et al. mTOR cross-talk in cancer and potential for combination therapy. Cancers 10, 23 (2018).
Felkai, L. et al. Characterization of mTOR activity and metabolic profile in pediatric rhabdomyosarcoma. Cancers 12, 1947 (2020).
Bang, H. et al. Correlation between RICTOR overexpression and amplification in advanced solid tumors. Pathol. Res. Pract. 216, 152734 (2020).
Bedics, G. et al. Germline MUTYH mutations and high-grade gliomas: Novel evidence for a potential association. Genes Chromosomes Cancer 61, 622–628 (2022).
Gieszer, B. et al. EGFR variant allele frequency predicts EGFR-TKI efficacy in lung adenocarcinoma: A multicenter study. Transl. Lung Cancer Res. 10, 662–674 (2021).
Krencz, I. et al. Metabolic adaptation as potential target in papillary renal cell carcinomas based on their in situ metabolic characteristics. Int. J. Mol. Sci. 23, 10587 (2022).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185, 563–575 (2022).
Chattopadhyay, A. et al. CNVIntegrate: The first multi-ethnic database for identifying copy number variations associated with cancer. Database 2021, 1–12 (2021).
Petővári, G. et al. In situ metabolic characterisation of breast cancer and its potential impact on therapy. Cancers 12, 2492 (2020).
Morrison Joly, M. et al. Two distinct mTORC2-dependent pathways converge on Rac1 to drive breast cancer metastasis. Breast Cancer Res. 19, 74 (2017).
Werfel, T. A. et al. Selective mTORC2 inhibitor therapeutically blocks breast cancer cell growth and survival. Cancer Res. 78, 1845–1858 (2018).
Alvarenga, A. W. et al. Evaluation of Akt and RICTOR expression levels in astrocytomas of all grades. J. Histochem. Cytochem. 65, 93–103 (2017).
Liu, Y. et al. mTORC2/Rac1 pathway predisposes cancer aggressiveness in IDH1-mutated glioma. Cancers 12, 787 (2020).
Petővári, G. et al. Targeting cellular metabolism using rapamycin and/or doxycycline enhances anti-tumour effects in human glioma cells. Cancer Cell. Int. 18, 211 (2018).
Akgül, S. et al. Opposing tumor-promoting and -suppressive functions of Rictor/mTORC2 signaling in adult glioma and pediatric SHH medulloblastoma. Cell Rep. 24, 463–478 (2018).
Wang, L. et al. Overexpression of Rictor protein in colorectal cancer is correlated with tumor progression and prognosis. Oncol. Lett. 14, 6198–6202 (2017).
Sticz, T. et al. mTOR activity and its prognostic significance in human colorectal carcinoma depending on C1 and C2 complex-related protein expression. J. Clin. Pathol. 70, 410–416 (2017).
Wen, F. F. et al. Expression of Raptor and Rictor and their relationships with angiogenesis in colorectal cancer. Neoplasma 67, 501–508 (2020).
Gulhati, P. et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 71, 3246–3256 (2011).
Javle, M. et al. Molecular characterization of gallbladder cancer using somatic mutation profiling. Hum. Pathol. 45, 701–708 (2014).
Kim, S. T. et al. Rapamycin-insensitive companion of mTOR (RICTOR) amplification defines a subset of advanced gastric cancer and is sensitive to AZD2014-mediated mTORC1/2 inhibition. Ann. Oncol. 28, 547–554 (2017).
Schmidt, K. M. et al. Inhibition of mTORC2 component RICTOR impairs tumor growth in pancreatic cancer models. Oncotarget 8, 24491–24505 (2017).
Wen, S. Y. et al. Rictor is an independent prognostic factor for endometrial carcinoma. Int. J. Clin. Exp. Pathol. 7, 2068–2078 (2014).
Kondo, S. et al. Raptor and rictor expression in patients with human papillomavirus-related oropharyngeal squamous cell carcinoma. BMC Cancer 21, 87 (2021).
Ruicci, K. M. et al. Disruption of the RICTOR/mTORC2 complex enhances the response of head and neck squamous cell carcinoma cells to PI3K inhibition. Mol. Oncol. 13, 2160–2177 (2019).
Naruse, T. et al. Therapeutic implication of mTORC2 in oral squamous cell carcinoma. Oral Oncol. 65, 23–32 (2017).
Gibault, L. et al. From PTEN loss of expression to RICTOR role in smooth muscle differentiation: Complex involvement of the mTOR pathway in leiomyosarcomas and pleomorphic sarcomas. Mod. Pathol. 25, 197–211 (2012).
Okada, T. et al. Integrin-α10 dependency identifies RAC and RICTOR as therapeutic targets in high-grade myxofibrosarcoma. Cancer Discov. 6, 1148–1165 (2016).
Krencz, I. et al. Expression of mTORC1/2-related proteins in primary and brain metastatic lung adenocarcinoma. Hum. Pathol. 62, 66–73 (2017).
Xu, Y. et al. Predictive values of genomic variation, tumor mutational burden, and PD-L1 expression in advanced lung squamous cell carcinoma treated with immunotherapy. Transl. Lung Cancer Res. 9, 2367–2379 (2020).
Cappello, F. et al. FFPE-based NGS approaches into clinical practice: The limits of glory from a pathologist viewpoint. J. Pers. Med. 12, 750 (2022).
Ramaiah, M. J. & Kumar, K. R. mTOR-Rictor-EGFR axis in oncogenesis and diagnosis of glioblastoma multiforme. Mol. Biol. Rep. 48, 4813–4835 (2021).
Jhanwar-Uniyal, M., Dominguez, J. F., Mohan, A. L., Tobias, M. E. & Gandhi, C. D. Disentangling the signaling pathways of mTOR complexes, mTORC1 and mTORC2, as a therapeutic target in glioblastoma. Adv. Biol. Regul. 83, 100854 (2022).
Li, X. et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 7, 33440–33450 (2016).
Miricescu, D. et al. PI3K/AKT/mTOR signaling pathway in breast cancer: From molecular landscape to clinical aspects. Int. J. Mol. Sci. 22, 173 (2020).
Moritz, S., Krause, M., Schlatter, J., Cordes, N. & Vehlow, A. Lamellipodin-RICTOR signaling mediates glioblastoma cell invasion and radiosensitivity downstream of EGFR. Cancers 13, 5337 (2021).
Gatti, G. et al. Role of Lamin A/C as candidate biomarker of aggressiveness and tumorigenicity in glioblastoma multiforme. Biomedicines 9, 1343 (2021).
Benavides-Serrato, A. et al. Specific blockade of Rictor-mTOR association inhibits mTORC2 activity and is cytotoxic in glioblastoma. PLoS ONE 12, e0176599 (2017).
Li, Y. et al. Murine models of IDH-wild-type glioblastoma exhibit spatial segregation of tumor initiation and manifestation during evolution. Nat. Commun. 11, 3669 (2020).
Petővári, G. et al. Inhibition of metabolic shift can decrease therapy resistance in human high-grade glioma cells. Pathol. Oncol. Res. 26, 23–33 (2020).
Roa, I. et al. Inactivation of tumor suppressor gene pten in early and advanced gallbladder cancer. Diagn. Pathol. 10, 148 (2015).
Simbolo, M. et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 5, 2839–2852 (2014).
Subbannayya, T. et al. PIM1 kinase promotes gallbladder cancer cell proliferation via inhibition of proline-rich Akt substrate of 40 kDa (PRAS40). J. Cell Commun. Signal. 13, 163–177 (2019).
Voss, M. H. et al. Phase 1 study of mTORC1/2 inhibitor sapanisertib (TAK-228) in advanced solid tumours, with an expansion phase in renal, endometrial or bladder cancer. Br. J. Cancer 123, 1590–1598 (2020).
Ishii, T. et al. Activation of the Akt-mTOR and MAPK pathways in dedifferentiated liposarcomas. Tumour Biol. 37, 4767–4776 (2016).
Kawasaki, G., Naruse, T., Furukawa, K. & Umeda, M. mTORC1 and mTORC2 expression levels in oral squamous cell carcinoma: An immunohistochemical and clinicopathological study. Anticancer Res. 38, 1623–1628 (2018).
Beck, J. T., Ismail, A. & Tolomeo, C. Targeting the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway: An emerging treatment strategy for squamous cell lung carcinoma. Cancer Treat. Rev. 40, 980–989 (2014).
Iksen, Pothongsrisit, S. & Pongrakhananon, V. Targeting the PI3K/AKT/mTOR signaling pathway in lung cancer: An update regarding potential drugs and natural products. Molecules 26, 4100 (2021).
Djukom, C. et al. Dual inhibition of PI3K and mTOR signaling pathways decreases human pancreatic neuroendocrine tumor metastatic progression. Pancreas 43, 88–92 (2014).
Zanini, S., Renzi, S., Giovinazzo, F. & Bermano, G. mTOR pathway in gastroenteropancreatic neuroendocrine tumor (GEP-NETs). Front. Endocrinol. 11, 562505 (2020).
Mpilla, G. B. et al. PAK4-NAMPT dual inhibition sensitizes pancreatic neuroendocrine tumors to everolimus. Mol. Cancer Ther. 20, 1836–1845 (2021).
Vandamme, T. et al. Long-term acquired everolimus resistance in pancreatic neuroendocrine tumours can be overcome with novel PI3K-AKT-mTOR inhibitors. Br. J. Cancer 114, 650–658 (2016).
Bobiński, M. et al. Gemcitabine and selected mTOR inhibitors in uterine sarcomas and carcinosarcoma cells—An isobolographic analysis. Int. J. Med. Sci. 17, 2987–2997 (2020).
Rubinstein, M. M. et al. Phase 2 study of LY3023414 in patients with advanced endometrial cancer harboring activating mutations in the PI3K pathway. Cancer 126, 1274–1282 (2020).
Guan, B. et al. Tumor-suppressive microRNA-218 inhibits tumor angiogenesis via targeting the mTOR component RICTOR in prostate cancer. Oncotarget 8, 8162–8172 (2017).
Micevic, G. et al. DNMT3b modulates melanoma growth by controlling levels of mTORC2 component RICTOR. Cell Rep. 14, 2180–2192 (2016).
Oneyama, C. et al. MiR-424/503-mediated Rictor upregulation promotes tumor progression. PLoS ONE 8, e80300 (2013).
Uesugi, A. et al. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 71, 5765–5778 (2011).
Zheng, G. et al. The miR-491-3p/mTORC2/FOXO1 regulatory loop modulates chemo-sensitivity in human tongue cancer. Oncotarget 6, 6931–6943 (2015).
Scattone, A. et al. Discordance between FISH, IHC, and NGS analysis of ALK status in advanced non-small cell lung cancer (NSCLC): A brief report of 7 Cases. Transl. Oncol. 12, 389–395 (2019).
Pauletti, G. et al. Assessment of methods for tissue-based detection of the HER-2/neu alteration in human breast cancer: A direct comparison of fluorescence in situ hybridization and immunohistochemistry. J. Clin. Oncol. 18, 3651–3664 (2000).
Dybdal, N., Leiberman, G. & Anderson, S. Determination of HER2 gene amplification by fluorescence in situ hybridization and concordance with the clinical trials immunohistochemical assay in women with metastatic breast cancer evaluated for treatment with trastuzumab. Breast Cancer Res. Treat. 93, 3–11 (2005).
Mass, R. D. et al. Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin. Breast Cancer 6, 240–246 (2005).
Sakre, N. et al. RICTOR amplification identifies a subgroup in small cell lung cancer and predicts response to drugs targeting mTOR. Oncotarget 8, 5992–6002 (2017).
Mao, B. et al. Overview of research into mTOR inhibitors. Molecules 27, 5295 (2022).
Ali, E. S. et al. Recent advances and limitations of mTOR inhibitors in the treatment of cancer. Cancer Cell Int. 22, 284 (2022).
Park, S. et al. Biomarker-driven phase 2 umbrella trial study for patients with recurrent small cell lung cancer failing platinum-based chemotherapy. Cancer 126, 4002–4012 (2020).
Acknowledgements
The authors are grateful to Dóra Kapczár, Erik Zajta, Lajos Hegyi and Csilla Kurucz for their excellent technical assistance at the Department of Pathology and Experimental Cancer Research, Semmelweis University (Budapest, Hungary). The authors thank Bálint Scheich for his help in classifying CNS tumours and Catherine Cervi for editing and peer-reviewing this manuscript. They would also like to thank András Khoór at the Department of Laboratory Medicine and Pathology, Mayo Clinic (Jacksonville, FL, USA) for initiating collaborations with us, research activity and help regarding RICTOR amplification studies of lung cancers.
Funding
Recent research at the Department of Pathology and Experimental Cancer Research (Semmelweis University, Budapest, Hungary) has been funded by the National Research, Development and Innovation Office of Hungary (NKFIH); TKP2021-EGA-24, TKP2021-NVA-15, NKFI-FK-128404, NKFI-K-142799. Our research is also supported by EFOP-3.6.3-VEKOP-16-2017-00009 (T.D.; D.M.; D.S.), ÚNKP-22-4-I-SE-12 (T.D.) and the Stephen W. Kuffler Research Grant (T.D.), H2020 (SGA No. 739593) and Elixir Hungary.
Author information
Authors and Affiliations
Contributions
A.S. designed this research work and supervised the literature review and manuscript preparation. All authors contributed to performing experimental and/or molecular measurements. G.B. helped to collect NGS results from the institutional diagnostic database. D.S. performed FISH, ddPCR and IHC staining. Á.N. and N.N. supervised ddPCR establishment, and G.P. helped with the evaluation of FISH analyses. Under the supervision of C.B., molecular analyses were performed. I.K., A.R., J.P., K.D., A.T. and Z.S. helped in the evaluation of IHC, clinical diagnoses and collection of patient material. Results were compiled by D.S., T.D. and D.M. and contributed to the writing of this manuscript. All authors read and carefully revised the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sztankovics, D., Krencz, I., Moldvai, D. et al. Novel RICTOR amplification harbouring entities: FISH validation of RICTOR amplification in tumour tissue after next-generation sequencing. Sci Rep 13, 19610 (2023). https://doi.org/10.1038/s41598-023-46927-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-46927-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.