Abstract
Breast cancer (BRCA) is known as the leading cause of death in women worldwide and has a poor prognosis. Traditional therapeutic strategies such as surgical resection, radiotherapy and chemotherapy can cause adverse reactions such as drug resistance. Immunotherapy, a new treatment approach with fewer side effects and stronger universality, can prolong the survival of BRCA patients and even achieve clinical cure. However, due to population heterogeneity and other reasons, there are still certain factors that limit the efficacy of immunotherapy. Therefore, the importance of finding new tumor immune biomarker cannot be emphasized enough. Studies have reported that LGALS2 was closely related to immunotherapy efficacy, however, it is unclear whether it can act as an immune checkpoint for BRCA immunotherapy. In the current study, changes in LGALS2 expression were analyzed in public datasets such as TCGA-BRCA. We found that LGALS2 expression was associated with immune infiltration, drug resistance and other characteristics of BRCA. Moreover, high LGALS2 expression was closely related to immunotherapy response, and was associated with methylation modifications and clinical resistance for the first time. These findings may help to elucidate the role of LGALS2 in BRCA for the development and clinical application of future immunotherapy strategies against BRCA.
Similar content being viewed by others
Introduction
There is no doubt that cancer remains one of the leading causes of death in most countries around the world1,2. According to GLOBOCAN 2020 data, there are about 2.3 million new cases of breast cancer (BRCA) in women worldwide, accounting for 11.7% of all malignant tumors3. BRCA has become the most prominent cause of mortality in women worldwide, with new cancer cases projected to increase from 24% in 2018 to more than 46% by 20404,5. Triple negative breast cancer (TNBC) is a type of BRCA without the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). Compared with other subtypes, TNBC is more prone to recurrence and metastasis, and has a lower survival rate6,7. It is worth noting that after receiving conventional treatments such as surgical resection, radiotherapy, and chemotherapy, there are still some patients who fail to benefit from the treatment8,9. In addition, the emergence of drug resistance and other phenomena during the treatment process also reduce the efficacy of the clinical treatments. Therefore, it is crucial to find a useful biomarker that can be used for clinical diagnosis/prognosis, with a potential for reducing clinical drug resistance.
Immunotherapy refers to a treatment method that artificially enhances and/or rebuilds the immune system to prevent and resist infection when the patient's body is in a low or hyperactive immune state10,11. Studies have demonstrated the therapeutic effects of immunotherapy in BRCA, STAD, LIHC, GBM and other cancer types7,12,13,14,15. The current immunotherapy regimen for tumors mainly includes monoclonal antibodies, tumor vaccines and non-specific immunotherapy, the purpose of which is to balance the immune system, so that cancer cells are eliminated without inducing autoimmune inflammation16,17. In addition, the abnormal expression of CTLA4, TIM-3, PD-1, TIGIT, HVEM and other immune checkpoint molecules is associated with many diseases. Immunotherapy with check point inhibitors increases the aggressiveness of the host's immune system against tumor cells by inhibiting the binding of programmed death receptors and their ligands18,19,20,21. At present, a large number of immune checkpoints and T cells are in the clinical and preclinical development stages, providing new reference for future tumor treatment strategies22,23,24.
Galectin-2 (LGALS2) is a homodimer consisting of 130 amino acids and a member of the galectin family25. They are known to bind to β-galactoside and contain at least one carbohydrate recognition domain (CRD), which plays an important role in many physiological and pathological processes such as cell adhesion, apoptosis, inflammatory response, and tumor metastasis26,27,28. There are currently 11 known human galectins and 15 known animal galectins29,30. Galectin-1, -3, -7, -8 and -9 are closely related to tumor immune escape31. LGALS2 is also closely related to immunotherapy response and plays an active role in cancer therapy32,33,34.
Based on public databases such as TCGA and METABRI, this study analyzed the expression levels of LGALS2 and the clinicopathological characteristics, diagnosis and prognosis, immune infiltration, etc. of BRCA patients. The results showed that LGALS2 was lowly expressed in BRCA patients (P < 0.001), and patients with higher LGALS2 expression had longer survival time (P = 0.014), especially LGALS2 had a better diagnostic potential in TNBC patients (AUC = 0.787). Analyses related to immune infiltration, GO and KEGG analysis, GSVA, enrichment analysis, etc. indicated that LGALS2 participated in the immune response of BRCA. Single-cell sequencing further demonstrated that LGALS2 was specifically highly expressed in T cells and could serve as a biomarker for immunotherapy response in BRCA patients.
At the same time, LGALS2 mRNA expression was negatively correlated with LGALS2 promoter methylation level and DNA methyltransferase expression level, and LGALS2 was associated with reduced IC50 values of several clinically used anticancer drugs. In conclusion, LGALS2 and BRCA have potential immunotherapeutic value. Therefore, regulating LGALS2 may be a novel strategy for the treatment of BRCA patients and LGALS2 might be a novel biomarker for BRCA immunotherapy.
Materials and methods
BRCA datasets
The Cancer Genome Atlas (TCGA), the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), the Gene Expression Omnibus (GEO), the Cancer Cell Line Encyclopedia (CCLE), the cBioPortal database and Connectivity Map (CMap) were used to obtain gene expression and pertinent prognostic and clinicopathological data for BRCA patients. In addition, all data were downloaded from public databases and analyzed using the R (version 4.0.3) and R Bioconductor.
Functional enrichment analysis
In the TCGA dataset (https://portal.gdc.com), we downloaded RNA-sequencing expression (level 3) profiles and clinical information related to BRCA patients. According to the expression level of LGALS2 gene in the TCGA-BRCA dataset, they were divided into two groups: high (n = 551) and low (n = 550). The R package Limma was used to study the differentially expressed mRNAs. Additionally, Adjusted P < 0.05 and Log2 (Fold Change) > 1 or < − 1 was defined as the threshold for the differential expression of mRNAs. We analyzed Gene Ontology (GO) function of the underlying mRNAs and enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway by using the ClusterProfiler package in R35.
Gene set variation analysis (GSVA)
We obtained the immune gene list from the Gene Set Enrichment Analysis (GSEA) (http://www.gsea-msigdb.org/). We calculated each BRCA sample's functional enrichment score using default parameters in R. With the pheatmap package in R, we mapped the enrichment results on a heatmap. Pearson correlation was used to determine the correlation between LGALS2 expression and immune responses.
Analysis of single‐cell clusters
GSE161529 was obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and processed using the Seurat package in R36. Genes expressed in more than three cells were considered as expressed, and each cell had to express 200 genes. The FindVariableFeatures function was used to identify the most variable genes from raw UMI counts. The variable genes were used in Principal components analysis (PCA). With a resolution of 0.6, the function FindClusters revealed shared nearest neighbor based on PCA using the first 20 principal components. Two-dimensional representations of the cell states were obtained using Uniform Manifold Approximation and Projection (UMAP) dimensional reduction analysis. Based on the CellMarker database (http://xteam.xbio.top/CellMarker/) and existing literature, the significant genes were used to assign cluster identity to the cell types37.
Correlation analysis of methylation expression
In the TCGA dataset (https://portal.gdc.com), we downloaded RNA-sequencing expression (level 3) profiles and illumina human methylation 450 states related to BRCA. In this study, differentially expressed mRNAs were visualized using the R package ggplot, and the data was transformed to Log2 (Fold Change)38.
Correlation analysis of IC50
In the TCGA dataset (https://portal.gdc.com), we downloaded RNA-sequencing expression (level 3) profiles and clinical information related to BRCA. Using Genomics of Drug Sensitivity in Cancer (GDSC), the chemotherapeutic response was predicted for each sample using the pRRophetic package in R. An estimation of the half-maximal inhibitory concentrations (IC50) was performed by using ridge regression with all parameters set to their default values39.
Small molecule targeted drugs screening of LGALS2 in BRCA
The Connectivity Map (CMap) (http://www.broad.mit.edu/cmap) was used to search for potential small molecule targeted drugs. Those small molecule drugs with |score|> 0.2 and P < 0.05 were recognized as the potential therapeutic drugs targeting LGALS2 in BRCA40,41.
Statistical analyses
All statistical analyses were performed using the R (version 4.0.3) software. Kaplan–Meier survival curves were constructed and compared with log-rank tests. The Spearman correlation analysis was used to examine correlations among variables without normal distributions. Data were analyzed between two and multiple groups by Student's t test and one-way ANOVA. P < 0.05 was considered to be statistically significant. (ns, P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Results
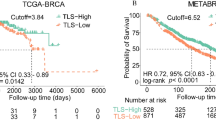
LGALS2 expression associates with diagnosis and prognosis in BRCA patients
The TCGA database was used to analyze the expression of LGALS2 in various normal and tumor tissues. The results showed that LGALS2 expression was statistically significant among 11 groups including BLCA (P < 0.01) (Fig. 1A), and was much lower in tumor groups, especially in BRCA (P < 0.001) (Fig. 1B). In addition, the expression levels of LGALS2 in different tissues (n = 29) and different human breast cancer cell lines (n = 68) were also analyzed based on the CCLE database, which was found to be consistent with the TCGA database results (Fig. S1A, B). Meanwhile, the above cell lines were classified into subtypes and our analyses showed no significant differences between these groups (P > 0.05) (Fig. S1C)42,43,44. And then, we further analyzed the diagnostic and prognostic value of LGALS2 in BRCA. The Kaplan–Meier (KM) curve indicated that BRCA patients with high LGALS2 expression had better survival rates (P = 0.014) (Fig. 1C), also in ER, HER2 and PR patients (P < 0.01) (Fig. 1E–G). Moreover, the receiver-operating characteristic (ROC) curve showed the diagnostic value of LGALS2 in BRCA patients (AUC = 0.601) (Fig. 1D) and their different subtypes (Fig. 1H–J). implying the potential value of LGALS2 in the diagnosis and prognosis of BRCA.
Transcriptional alterations and diagnostic/prognostic value of LGALS2 in BRCA patients. (A) Expression of LGALS2 in paracancers and tumors in TCGA database. The significance of the difference was tested with an unpaired student’s t test. (B) LGALS2 expression in BRCA patients based on the TCGA database. The significance of the difference was tested with an unpaired student’s t test. (C) KM analysis of the diagnostic value of LGALS2 in TCGA database. (D) ROC analysis of the prognostic value of LGALS2 in TCGA database. (E–G) KM analysis of overall survival of LGALS2 in patients with different subtypes of BRCA. (H–J) ROC analysis of prognostic of LGALS2 in patients with different subtypes of BRCA. ns, P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Analysis of various clinical factors associated with LGALS2 expression
The clinical patients with different expression of LGALS2 showed the distinct patterns of clinical and pathological characteristics. The change in LGALS2 expression with chemotherapy or radiotherapy, age and gender, pathologic-stage, T-stage, N-stage, M-stage and survival status are shown in Fig. 2A. The expression of LGALS2 was not statistically significant in groups with pathological-, T-, N- or M-stage, radiotherapy, drug treatment (Fig. 2B–G) and gender (Fig. 2I), but except for the age group (P < 0.01) (Fig. 2H). The clinical factors associated with LGALS2 expression (e.g., AJCC-stage, age, gender, race, etc.) were also extracted from the cBipPortal database to complement the results of the TCGA database (Fig. S2)45. Consistent with the prior results, LGALS2 expression levels were found to significantly differ only with patient diagnosis age (P < 0.05) and race (P < 0.001). The results of this study did not reveal a statistically significant difference based on the clinical stage.
Analysis of various clinical factors associated with LGALS2 expression. (A) The landscape of LGALS2-related clinicopathological features of BRCA in TCGA database. (B–I) LGALS2 and various clinicopathological features of BRCA in TCGA databases. (B–D) LGALS2 was not significantly different in Pathologic-, T-, N-stage in TCGA database. The significance of the difference was tested by one‐way ANOVA. (E–I) LGALS2 was not significantly different in M-stage, radiation therapy, pharmaceutical therapy and gender in TCGA database. The significance of the difference was tested with an unpaired student’s t test. (H) there was a significant difference between LGALS2 expression and patient age in TCGA databases. The significance of the difference was tested with an unpaired student’s t test.
LGALS2 has diagnostic value for TNBC
Studies have reported that LGALS2 could be used as a therapeutic target for TNBC33. However, there are still some data limitations in the TCGA database, thus, the METABRIC database was used to analyze the expression of LGALS2 in the BRCA subtypes. When molecular markers were detected in various types of BRCA, the expression level of LGALS2 was significantly elevated in ER-negative, HER2-negative, and PR-negative groups (P < 0.0001) (Fig. 3A–C). We then investigated LGALS2 distribution in different subtypes defined by the METABRIC database. Results showed that LGALS2 was significantly enriched in the Claudin-low subtype compared to the other subtypes (P < 0.0001) (Fig. 3D). LGALS2 expression specificity was assessed using ROC curves. The area under the curve (AUC) was up to 78.7% in the METABRIC database (P < 0.0001) (Fig. 3E). It appeared that LGALS2 was significantly enriched in TNBC, suggesting its diagnostic potential as a biomarker and providing a reference for the clinical diagnosis of TNBC.
Diagnostic analysis of LGALS2 expression with TNBC. (A–C) Expression of LGALS2 in ER, HER2, PR. LGALS2 was enriched in the negative subtype of BRCA in the Metabric database. The significance of the difference was tested with an unpaired t test. (D) LGALS2 was highly expressed in claudin-low subtype of BRCA in the Metabric database. The significance of the difference was tested by one‐way ANOVA. (E) The ROC curve showed the high‐expression specificity of LGALS2 in TNBC in the Metabric database.
LGALS2 regulates immune responses in a T cell-dependent manner
LGALS2 expression was investigated in the TCGA database using the LIMMA R package to explore its potential biological function in BRCA. It generated 300 genes, and 279 genes were upregulated, 21 genes were downregulated (|logFC|> 1, adjusted P < 0.05) (Fig. 4A,B). In the GO and KEGG analyses, LGALS2 functions were primarily related to cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, chemokine signaling pathway, T cell activation, leukocyte cell–cell adhesion, regulation of T cell activation, etc. (Fig. 4C–F). Based on these findings, combined with the previous results related to immune infiltration T cells and its association with LGALS2 expression, we speculated that LGALS2 might be closely associated with immunotherapy outcome and contribute to immune response in BRCA patients.
GO and KEGG analysis of LGALS2. (A) Volcano plot of differentially expressed genes. Red dots indicate upregulated genes, blue dots indicate downregulated genes, grey dots indicate not significant. (B) Heat map showing the differentially expressed genes, and the different colors represent the trend of gene expression. The top 50 up-regulated genes and top 50 down-regulated genes were shown in this figure (C,E) KEGG enrichment analysis. (D,F) GO enrichment analysis. Colors represent the significance of differential enrichment, the size of the circles represents the number of genes, the larger the circle, the greater the number of genes. (|LogFC|> 1, Adjusted P < 0.05).
LGALS2 expression positively correlates with T cell-mediated immune responses
Cancer cells die immunologically due to lymphocyte activation (including NK cells, T cells, and B cells) and the release of chemokines and cytokines46,47. Therefore, we investigated the effects of LGALS2 expression on immune pathways and cytokines. TCGA gene set variation analysis (GSVA) was used to determine the immune process enrichment score. Based on correlation analysis between LGALS2 expression and enrichment score, LGALS2 expression was positively correlated with most immune functions, but not with activin receptor signaling pathway and plasma membrane organization (Fig. 5A). LGALS2 expression was associated with various immune cells as shown in Fig. 5B, and was most closely associated with T cells. Moreover, the immune infiltration data showed a higher degree of enrichment of LGALS2 and T cells (P < 0.001, r = 0.737) (Fig. 5C). The above results indicated that LGALS2 expression was associated with T cell activity and immunotherapy response in BRCA patients.
Correlation analysis between LGALS2 and immune function. (A) Heatmap showing LGALS2 expression and the enrichment scores of immune functions of each patient in TCGA, and the column and line graph on the right shows the R and P values for correlation analysis. (B) Analysis of the correlation between LGALS2 and immune cells. The size of the circle represents the correlation, with the larger the circle the higher the correlation. (C) Scatter plot of LGALS2 and T cell enrichment. LGALS2 expression levels were positively correlated with T cell enrichment. The significance of the difference was tested by Spearman correlation analysis.
LGALS2 positively correlates with established cancer immune checkpoints
In previous studies, LGALS2 was shown to be involved in tumor immunity48, thus we investigated the relationship between LGALS2 expression in the TCGA dataset and immune checkpoints, including CD200R1, CD47, CTLA4, TIM-3, PD-1, TIGIT, HVEM and CD69. LGALS2 expression showed a strong relationship with these immune checkpoints18,19,49 (Fig. 6A). Additionally, there was a positive correlation between LGALS2 expression and inflammatory-related metagenes (including GATA3, C3, CD44, C5, CEBPB, CD4, CCL11, AKT1, CD2)50,51,52,53,54 (Fig. 6B). Furthermore, these results indicated that LGALS2 expression was associated with tumor immunotherapy and could serve as a biomarker for response to immunotherapy.
Analysis of the correlation between LGALS2 and T cell immunity and inflammation. (A) Pearson correlation between LGALS2 and inhibitory immune checkpoints. The color of the band represented the P‐value. The correlation was tested by Pearson correlation analysis. (B) Correlation matrix of LGALS2 and inflammatory‐related metagenes. The bottom left showed the correlation coefficient. which are shown as a scale of the pie charts. The correlation was tested by Pearson correlation analysis.
T cells express high levels of LGALS2
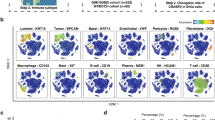
We analyzed public datasets from the GEO database using R for single-cell sequencing. UMAP dimensionality reduction analysis was used to obtain a two-dimensional representation of cell state and 12 clusters were created (Fig. 7A). We found that LGALS2 was mainly enriched and highly expressed in the clusters 4 and 5 (Fig. 7B). Cellular markers enabled us to identify the cluster 4 and cluster 5. Based on the expression of multiple cellular markers, such as CD4, CD68, CD83 and so on55,56,57, clusters 4 and 5 were categorized as T cells (Fig. 7C,D).
Single-cell sequencing analysis of LGALS2. (A) Single-cell sequencing analysis showing the cellular subtypes of BRCA, based on the GEO database (GSE161529). (B) LGALS2 is highly expressed in cluster 4 and 5. (C,D) Expression of T cell markers in different subtypes. Verification of cluster 4 and cluster 5 as T cells.
In the subsequent analyses, we showed that LGALS2 expression was closely associated with the immune system and interacted with T cells. Thus, upregulating LGALS2 expression was associated with the activation and differentiation of T cells, induction of immune responses and promotion of the necrosis or apoptosis of breast cancer cells.
LGALS2 expression level positively correlates with methylation modification
In light of the importance of DNA methylation in regulating gene expression58,59, we examined LGALS2-associated methylation level. A significant increase in promoter methylation of LGALS2 was observed in tumor samples as compared to the normal group (P < 0.0001) (Fig. 8A), and this was also significantly associated with the tumor type and patient age (Fig. 8B,C), which were inversely correlated with LGALS2 mRNA expression level (Fig. 1B). At the same time, the expression levels of DNA methyltransferases (DNMT) (including DNMT1, DNMT3A and DNMT3B) in the normal and tumor samples from TCGA database were also determined. The results showed that DNMT was highly expressed in breast cancer (P < 0.0001) (Fig. 8D–F). Furthermore, the results suggested that methylation occurred 2400–2700 bp downstream of LGALS2 transcription start sites (cg23835646, cg11081833 and cg26651950) (P < 0.001) (Fig. 8G–I). Due to this, DNMT might affect the transcription process by adding methyl groups to LGALS2 without affecting its sequence, thereby reducing the expression of LGALS2, and the methylation modification of LGALS2 might be one of the reasons for its decreased mRNA expression.
Methylation-related analysis of LGALS2. (A) Promoter methylation level of LGALS2 in BRCA. The significance of the difference was tested with an unpaired student’s t test. (B) LGALS2 promoter methylation expression based on BRCA subclasses. The significance of the difference was tested with an unpaired student’s t test. (C) LGALS2 promoter methylation expression based on age of BRCA patients. The significance of the difference was tested with an unpaired student’s t test. (D–F) Relationship between DNA methyltransferases (DNMT1, DNMT3A and DNMT3B) and LGALS2 expression in BRCA patients. The significance of the difference was tested with an unpaired student’s t test. (G–I) Methylation probe linked to LGALS2 in BRCA patients. The significance of the difference was tested by Spearman correlation analysis. ns, P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
LGALS2 expression associates with clinical sensitivity to anticancer drugs
To further explore LGALS2 as a potential therapeutic target in BRCA, we performed CMap analysis and obtained 15 compounds with positive and negative correlations, respectively (Fig. S4A)60,61. It has been reported that cancer patients often suffer from drug resistance, which leads to relapses and reduced survival rates62. The role of LGALS2 expression in breast cancer drug resistance was therefore studied. Surprisingly, we found a negative correlation between LGALS2 expression and resistance in BRCA (P < 0.0001) (Fig. S3). High levels of LGALS2 were associated with a significant reduction in the IC50 values of several clinical anticancer drugs (including camptothecin, parthenolide and paclitaxel)63,64,65,66,67,68,69,70,71 (Fig. 9). Meanwhile, CMap analysis further evaluated to validate the above drugs. The results also showed that these drugs were significantly associated with LGALS2 expression in BRCA (P < 0.01) (Fig. S4B–E). Therefore, LGALS2 expressed levels might be associated with the increased sensitivity of cancer cells to clinical drugs and longer survival of cancer patients.
Discussion
Breast cancer is the number most common cause of cancer related mortality in women globally, and TNBC accounts for about 10% to 15% of that72. Although the patient's condition is improved after surgical resection, radiotherapy, chemotherapy and other therapeutic interventions, adverse reactions such as metastasis and drug resistance contribute to the overall unsatisfactory treatment outcomes73,74. Compared with other therapeutic strategies, cancer immunotherapy causes fewer side effects and is more universal, which prolongs the survival time of patients and even achieves clinical cure, and thus is a promising treatment strategy75,76. However, there are still some limitations such as tumor immune escape, tumor mutation burden, and adaptive immune resistance, which suppress the therapeutic potential of cancer immunotherapy38,77. Therefore, it is crucial to find a new biomarker for evaluating immunotherapy response in breast cancer treatment.
In this study, we found that LGALS2 expression was low in breast cancer patients and was lower in the breast tissue relative to other tissues, and patients with high LGALS2 expression had a better prognosis (P = 0.014). Furthermore, we show that LGALS2 could be used as a diagnostic marker for breast cancer (AUC = 0.601), and has a better diagnostic value in TNBC (P < 0.0001). The ROC analysis showed that the AUC-value was 0.787. The above findings suggest that LGALS2 has a good prognostic value for BRCA patients, especially for TNBC patients.
However, in the analysis of LGALS2 and related clinicopathological factors based on the TCGA and cBioPortal database, there was no statistically significant correlation between LGALS2 expression and the commonly used clinical stages. Braud VM et al.78 reported that KLRB1 was a predictive marker for the survival status of patients, but was not related to the degree of tumor cell invasion, and our findings were consistent with them. In addition, the current common tumor staging is based on the length of the tumor, the status of the ipsilateral axillary lymph node, and the presence of distant metastasis79,80. Therefore, we speculated that LGALS2 might not be directly involved in the migration and development of breast cancer. Of course, data is an important factor affecting the results, including the data collection process, the size and quality of the data, the regional distribution of patients, and the heterogeneity and bias of the data81,82.
DNA methylation is often dysregulated during tumorigenesis and cancer progression, which are manifested by the reduction of the overall methylation level of the genome and the inactivation of the local DNA regions83,84. Whether the low expression of LGALS2 in breast cancer patients was related to DNA methylation modification has not yet been reported. Here, we found that LGALS2 promoter methylation levels were high in the tumor groups (P < 0.001) and were closely related to the tumor subtypes and patient age (P < 0.05). At the same time, DNA methyltransferases (DNMT) (including DNMT1, DNMT3A, DNMT3B) were also highly expressed in tumors (P < 0.001). Namely, LGALS2 expression was negatively correlated with LGALS2 promoter methylation and DNA methyltransferase. As a result, we speculated that DNMT regulated methylation modifications in the LGALS2 promoter region to inhibit the transcription of LGALS2, resulting in decreased LGALS2 expression. Moreover, the results suggested that methylation occurred 2400-2700 bp downstream of LGALS2 transcription start sites (cg23835646[TSS + 2561], cg11081833[TSS + 2683], cg26651950[TSS + 2445]) (P < 0.001). Therefore, reducing the methylation level of the LGALS2 promoter, thereby reversing the occurrence and development of breast cancer, could serve as a new strategy for breast cancer treatment. However, further research is warranted to verify the above findings.
Previous studies reported that LGALS2 induced T lymphocyte apoptosis, improved colitis, and prevented preeclampsia27,85. During cancer immunotherapy, T cells play a key role in the antitumor response and are closely associated with the effective inhibition of immune checkpoints86,87. In this study, GO and KEGG analysis, GSVA, enrichment score, etc. revealed that LGALS2 was associated with increased T cell infiltration (P < 0.001) and had a positive regulatory relationship with immune response. The data used in these analyses were all obtained from the average of multiple data samples, but we could not obtain information regarding inherent tumor heterogeneity. However, single-cell sequencing solves this problem well, with higher sensitivity and accuracy, and more reliable results. Cell markers such as CD4, CD68, AIF1, PLEK, and others were used to identify T cell clusters56,88, and we found LGALS2 to be specifically highly expressed in T cell clusters. The above findings suggested that LGALS2 regulated T cells to induce immune response in the body and participated in tumor immunotherapy.
The combination of immunotherapy and chemotherapy (so-called chemo-immunotherapy) has demonstrated excellent therapeutic effects in the clinical treatment of cancer patients89,90, but adverse reactions such as drug resistance significantly limit the therapeutic efficacy of the combination strategy91,92. The main cause of cancer recurrence is the development of multidrug resistance, which in turn is associated with increased expression of efflux transporters, epithelial-to-mesenchymal transformation and breast cancer stem cell resistance93. In addition, it has been demonstrated that the formation of an immunosuppressive niche increased the occurrence of drug resistance and suppressed the antitumor effects of T cells, reducing the efficacy of immunotherapy94. We therefore examined the role of LGALS2 in clinical drug resistance based on the GDSC database. The results showed that high LGALS2 expression was associated with a lower IC50 value of 3 clinical antitumor drugs, including camptothecin, which enhanced their drug sensitivity and reduced the occurrence of clinical drug resistance. Subsequently we also used CMap to analyse the correlation between these 3 drugs and LGALS2 in BRCA, and gratifyingly this result also showed that these drugs were significantly correlated with LGALS2 expression. Of course, the results of the database have to be verified by further wet experiments.
Twyman-Saint Victor et al.95 reported that the up-regulation of PD-L1 in melanoma cells was an important factor in the development of drug resistance and was related to T cell exhaustion. Interestingly, therapy- and resistance-related changes in T cells might constitute biomarkers of tumor response. Moreover, immune checkpoints such as PD-1/PD-L1, TIM-3, and TIGIT are known to play important roles in the treatment of NSCLC, OV, SKCM, BRCA, and have been recognized as biomarkers for immunotherapy response96,97. Through enrichment analysis and immune score, Jiang et al.98 identified that the expression level of STC2 was significantly positively correlated with the infiltration level of B cells, T cells and other immune cells in various cancer cells, and was significantly correlated with the sensitivity towards certain drugs, and suggested STC2 as a promising target for tumor immunotherapy. Therefore, LGALS2 could also serve as a potential marker for breast cancer immunotherapy.
Our study systematically and comprehensively validates that LGALS2 can be used as a diagnostic/prognostic marker for breast cancer, especially a good diagnostic marker for TNBC, and that it participates in tumor immunotherapy by regulating T cells. The result that high LGALS2 expression was associated with reduced occurrence of clinical tumor therapy resistance was even more intriguing for us. However, our study suffered from some limitations. First of all, this study solely relied on existing public data, thus necessitating further experimental evidence to verify and clarify the molecular mechanism of LGALS2 in BRCA. Secondly, the low expression of LGALS2 in BRCA was related to the methylation modification of its promoter region and the ability of LGALS2 to reduce the occurrence of clinical drug resistance was also identified for the first time in this study. However, further experimental and clinical studies are needed to confirm the above findings. Lastly, although the results of this study supported the involvement of LGALS2 in tumor immune regulation and immunotherapy, the underlying molecular mechanism needs further investigation.
Conclusion
In summary, our study shows that LGALS2 could be used as a diagnostic and prognostic marker for breast cancer, and that it regulates the biological activity of T cells to participate in tumor immunotherapy and reduces the occurrence of clinical drug resistance in patients. As a novel molecular biomarker for breast cancer treatment, LGALS2 may enable the development of novel immunotherapy strategies that are of a high clinical relevance in the future. The findings from this study need further validation through in vitro and in vivo experiments to confirm the functions of LGALS2 and unravel the underlying molecular mechanisms in breast cancer.
Data availability
The datasets analyzed for this study can be found in the TCGA-BRCA project (http://www.cancer.gov/tcga), METABRIC (https://www.mercuriolab.umassmed.edu/metabric), GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161529) and GDSC (https://www.cancerrxgene.org/), CCLE (https://sites.broadinstitute.org/ccle/), cBioPortal (https://www.cbioportal.org/), CMap (https://clue.io/).
Abbreviations
- AUC:
-
Area under the curve
- AJCC:
-
American joint committee on cancer
- BRCA:
-
Breast cancer
- CMap:
-
Connectivity map
- CCLE:
-
Cancer cell line encyclopedia
- CRD:
-
Carbohydrate recognition domain
- ER:
-
Estrogen receptor
- GDSC:
-
Genomics drug sensitivity in cancer
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- HER2:
-
Human epidermal growth factor receptor 2
- IC50:
-
Half-maximal inhibitory concentration
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- KM:
-
Kaplan–Meier
- METABRIC:
-
Molecular taxonomy of breast cancer international consortium
- PR:
-
Progesterone receptor
- ROC:
-
Receiver‐operating characteristic
- TCGA:
-
The cancer genome atlas
- THPA:
-
The human protein atlas
- TNBC:
-
Triple negative breast cancer
References
Bray, F., Laversanne, M., Weiderpass, E. & Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 127, 3029–3030. https://doi.org/10.1002/cncr.33587 (2021).
Xia, C. et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 135, 584–590. https://doi.org/10.1097/cm9.0000000000002108 (2022).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. https://doi.org/10.3322/caac.21492 (2018).
Heer, E. et al. Global burden and trends in premenopausal and postmenopausal breast cancer: A population-based study. Lancet Glob. Health 8, e1027–e1037. https://doi.org/10.1016/s2214-109x(20)30215-1 (2020).
Singh, D. D. & Yadav, D. K. TNBC: Potential targeting of multiple receptors for a therapeutic breakthrough, nanomedicine, and immunotherapy. Biomedicines https://doi.org/10.3390/biomedicines9080876 (2021).
Zhu, Y., Zhu, X., Tang, C., Guan, X. & Zhang, W. Progress and challenges of immunotherapy in triple-negative breast cancer. Biochim. Biophys. Acta Rev. Cancer 1876, 188593. https://doi.org/10.1016/j.bbcan.2021.188593 (2021).
Loibl, S., Poortmans, P., Morrow, M., Denkert, C. & Curigliano, G. Breast cancer. Lancet 397, 1750–1769. https://doi.org/10.1016/s0140-6736(20)32381-3 (2021).
Trayes, K. P. & Cokenakes, S. E. H. Breast cancer treatment. Am. Fam. Phys. 104, 171–178 (2021).
Lizée, G. et al. Harnessing the power of the immune system to target cancer. Annu. Rev. Med. 64, 71–90. https://doi.org/10.1146/annurev-med-112311-083918 (2013).
Riley, R. S., June, C. H., Langer, R. & Mitchell, M. J. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 18, 175–196. https://doi.org/10.1038/s41573-018-0006-z (2019).
Li, K., Zhang, A., Li, X., Zhang, H. & Zhao, L. Advances in clinical immunotherapy for gastric cancer. Biochim. Biophys. Acta Rev. Cancer 1876, 188615. https://doi.org/10.1016/j.bbcan.2021.188615 (2021).
De Martin, E. et al. Characterization of liver injury induced by cancer immunotherapy using immune checkpoint inhibitors. J. Hepatol. 68, 1181–1190. https://doi.org/10.1016/j.jhep.2018.01.033 (2018).
Emens, L. A. Breast cancer immunotherapy: Facts and hopes. Clin. Cancer Res. 24, 511–520. https://doi.org/10.1158/1078-0432.Ccr-16-3001 (2018).
Choi, B. D., Maus, M. V., June, C. H. & Sampson, J. H. Immunotherapy for glioblastoma: Adoptive T-cell strategies. Clin. Cancer Res. 25, 2042–2048. https://doi.org/10.1158/1078-0432.Ccr-18-1625 (2019).
Chen, D. S. & Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 39, 1–10. https://doi.org/10.1016/j.immuni.2013.07.012 (2013).
Brody, J., Kohrt, H., Marabelle, A. & Levy, R. Active and passive immunotherapy for lymphoma: Proving principles and improving results. J. Clin. Oncol. 29, 1864–1875. https://doi.org/10.1200/jco.2010.33.4623 (2011).
Su, Y. et al. Tumor-infiltrating T cells concurrently overexpress CD200R with immune checkpoints PD-1, CTLA-4, and TIM-3 in non-small-cell lung cancer. Pathobiology 88, 218–227. https://doi.org/10.1159/000511557 (2021).
Poh, S. L. & Linn, Y. C. Immune checkpoint inhibitors enhance cytotoxicity of cytokine-induced killer cells against human myeloid leukaemic blasts. Cancer Immunol. Immunother. 65, 525–536. https://doi.org/10.1007/s00262-016-1815-8 (2016).
Chauvin, J. M. & Zarour, H. M. TIGIT in cancer immunotherapy. J. Immunother. Cancer https://doi.org/10.1136/jitc-2020-000957 (2020).
Aubert, N., Brunel, S., Olive, D. & Marodon, G. Blockade of HVEM for prostate cancer immunotherapy in humanized mice. Cancers https://doi.org/10.3390/cancers13123009 (2021).
Kalbasi, A. & Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 20, 25–39. https://doi.org/10.1038/s41577-019-0218-4 (2020).
Heinhuis, K. M. et al. Enhancing antitumor response by combining immune checkpoint inhibitors with chemotherapy in solid tumors. Ann. Oncol. 30, 219–235. https://doi.org/10.1093/annonc/mdy551 (2019).
Curdy, N., Lanvin, O., Laurent, C., Fournié, J. J. & Franchini, D. M. Regulatory mechanisms of inhibitory immune checkpoint receptors expression. Trends Cell Biol. 29, 777–790. https://doi.org/10.1016/j.tcb.2019.07.002 (2019).
Sasaki, T. et al. Galectin-2 has bactericidal effects against Helicobacter pylori in a β-galactoside-dependent manner. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21082697 (2020).
Bänfer, S. & Jacob, R. Galectins. Curr. Biol. 32, R406-r408. https://doi.org/10.1016/j.cub.2022.03.065 (2022).
Meister, S. et al. Regulatory T cell apoptosis during preeclampsia may be prevented by Gal-2. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23031880 (2022).
Schlichtner, S. et al. Functional role of galectin-9 in directing human innate immune reactions to Gram-negative bacteria and T cell apoptosis. Int. Immunopharmacol. 100, 108155. https://doi.org/10.1016/j.intimp.2021.108155 (2021).
Manero-Rupérez, N., Martínez-Bosch, N., Barranco, L. E., Visa, L. & Navarro, P. The galectin family as molecular targets: Hopes for defeating pancreatic cancer. Cells https://doi.org/10.3390/cells9030689 (2020).
Salamanna, F., Veronesi, F., Frizziero, A. & Fini, M. Role and translational implication of galectins in arthritis pathophysiology and treatment: A systematic literature review. J. Cell. Physiol. 234, 1588–1605. https://doi.org/10.1002/jcp.27026 (2019).
Laderach, D. J. & Compagno, D. Unraveling how tumor-derived galectins contribute to anti-cancer immunity failure. Cancers https://doi.org/10.3390/cancers13184529 (2021).
Li, H., Zhao, L., Lau, Y. S., Zhang, C. & Han, R. Genome-wide CRISPR screen identifies LGALS2 as an oxidative stress-responsive gene with an inhibitory function on colon tumor growth. Oncogene 40, 177–188. https://doi.org/10.1038/s41388-020-01523-5 (2021).
Ji, P. et al. In vivo multidimensional CRISPR screens identify Lgals2 as an immunotherapy target in triple-negative breast cancer. Sci. Adv. 8, eabl8247. https://doi.org/10.1126/sciadv.abl8247 (2022).
Barrow, H. et al. Serum galectin-2, -4, and -8 are greatly increased in colon and breast cancer patients and promote cancer cell adhesion to blood vascular endothelium. Clin. Cancer Res. 17, 7035–7046. https://doi.org/10.1158/1078-0432.Ccr-11-1462 (2011).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. https://doi.org/10.1038/nbt.4096 (2018).
Zhang, X. et al. Cell marker: A manually curated resource of cell markers in human and mouse. Nucl. Acids Res. 47, D721-d728. https://doi.org/10.1093/nar/gky900 (2019).
Anuraga, G. et al. Potential prognostic biomarkers of NIMA (never in mitosis, Gene A)-related kinase (NEK) family members in breast cancer. J. Pers. Med. https://doi.org/10.3390/jpm11111089 (2021).
Jiang, Q. et al. Establishment of an immune cell infiltration score to help predict the prognosis and chemotherapy responsiveness of gastric cancer patients. Front. Oncol. 11, 650673. https://doi.org/10.3389/fonc.2021.650673 (2021).
Wu, X. et al. HOXB7 acts as an oncogenic biomarker in head and neck squamous cell carcinoma. Cancer Cell Int. 21, 393. https://doi.org/10.1186/s12935-021-02093-6 (2021).
Wang, C. Y. et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am. J. Cancer Res. 10, 95–113 (2020).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. https://doi.org/10.1038/nature11003 (2012).
Kao, T. J. et al. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging 13, 17970. https://doi.org/10.18632/aging.203345 (2021).
Li, W., Ma, J. A., Sheng, X. & Xiao, C. Screening of CXC chemokines in the microenvironment of ovarian cancer and the biological function of CXCL10. World J. Surg. Oncol. 19, 329. https://doi.org/10.1186/s12957-021-02440-x (2021).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. https://doi.org/10.1158/2159-8290.Cd-12-0095 (2012).
Oshi, M. et al. Immune cytolytic activity is associated with reduced intra-tumoral genetic heterogeneity and with better clinical outcomes in triple negative breast cancer. Am. J. Cancer Res. 11, 3628–3644 (2021).
Wang, Z. L. et al. Immune cytolytic activity is associated with genetic and clinical properties of glioma. Front. Immunol. 10, 1756. https://doi.org/10.3389/fimmu.2019.01756 (2019).
Niu, J. et al. The in vivo roles of galectin-2 from Nile tilapia (Oreochromis niloticus) in immune response against bacterial infection. Fish Shellfish Immunol. 106, 473–479. https://doi.org/10.1016/j.fsi.2020.08.011 (2020).
Liu, W. et al. HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160. J. Exp. Med. https://doi.org/10.1084/jem.20211112 (2021).
Stark, J. M., Tibbitt, C. A. & Coquet, J. M. The metabolic requirements of Th2 cell differentiation. Front. Immunol. 10, 2318. https://doi.org/10.3389/fimmu.2019.02318 (2019).
Jackson, W. D. et al. C3 drives inflammatory skin carcinogenesis independently of C5. J. Investig. Dermatol. 141, 404-414.e406. https://doi.org/10.1016/j.jid.2020.06.025 (2021).
Norris, P. A. A. et al. Anti-inflammatory activity of CD44 antibodies in murine immune thrombocytopenia is mediated by Fcγ receptor inhibition. Blood 137, 2114–2124. https://doi.org/10.1182/blood.2020009497 (2021).
Ma, J., Yang, X. & Chen, X. C/EBPβ is a key transcription factor of ox-LDL inducing THP-1 cells to release multiple pro-inflammatory cytokines. Inflamm. Res. 70, 1191–1199. https://doi.org/10.1007/s00011-021-01509-3 (2021).
Scabia, G. et al. Reduced ccl11/eotaxin mediates the beneficial effects of environmental stimulation on the aged hippocampus. Brain Behav. Immunity 98, 234–244. https://doi.org/10.1016/j.bbi.2021.08.222 (2021).
Schelker, M. et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat. Commun. 8, 2032. https://doi.org/10.1038/s41467-017-02289-3 (2017).
Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342-1356.e1316. https://doi.org/10.1016/j.cell.2017.05.035 (2017).
Szabolcs, M. J. et al. Apoptosis and increased expression of inducible nitric oxide synthase in human allograft rejection. Transplantation 65, 804–812. https://doi.org/10.1097/00007890-199803270-00007 (1998).
Zheng, F. et al. Expression and clinical prognostic value of m6A RNA methylation modification in breast cancer. Biomark. Res. 9, 28. https://doi.org/10.1186/s40364-021-00285-w (2021).
Nasir, A. et al. Nutrigenomics: Epigenetics and cancer prevention: A comprehensive review. Crit. Rev. Food Sci. Nutr. 60, 1375–1387. https://doi.org/10.1080/10408398.2019.1571480 (2020).
Lamb, J. et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. https://doi.org/10.1126/science.1132939 (2006).
Subramanian, A. et al. A next generation connectivity map: L1000 Platform and the first 1,000,000 profiles. Cell 171, 1437-1452.e1417. https://doi.org/10.1016/j.cell.2017.10.049 (2017).
Palomeras, S., Ruiz-Martínez, S. & Puig, T. Targeting breast cancer stem cells to overcome treatment resistance. Molecules https://doi.org/10.3390/molecules23092193 (2018).
Chiu, C. F. et al. The novel camptothecin derivative, CPT211, induces cell cycle arrest and apoptosis in models of human breast cancer. Biomed. Pharmacother. 128, 110309. https://doi.org/10.1016/j.biopha.2020.110309 (2020).
Wang, H. et al. Cisplatin prevents breast cancer metastasis through blocking early EMT and retards cancer growth together with paclitaxel. Theranostics 11, 2442–2459. https://doi.org/10.7150/thno.46460 (2021).
Giannone, G. et al. Oral etoposide in heavily pre-treated metastatic breast cancer: A retrospective series. Breast 38, 160–164. https://doi.org/10.1016/j.breast.2018.01.006 (2018).
Stovgaard, E. S. et al. The immune microenvironment and relation to outcome in patients with advanced breast cancer treated with docetaxel with or without gemcitabine. Oncoimmunology 10, 1924492. https://doi.org/10.1080/2162402x.2021.1924492 (2021).
Yu, K. D. et al. Effect of adjuvant paclitaxel and carboplatin on survival in women with triple-negative breast cancer: A phase 3 randomized clinical trial. JAMA Oncol. 6, 1390–1396. https://doi.org/10.1001/jamaoncol.2020.2965 (2020).
Berdan, C. A. et al. Parthenolide covalently targets and inhibits focal adhesion kinase in breast cancer cells. Cell Chem. Biol. 26, 1027-1035.e1022. https://doi.org/10.1016/j.chembiol.2019.03.016 (2019).
Zhou, Q. et al. Linc-ROR promotes the progression of breast cancer and decreases the sensitivity to rapamycin through miR-194-3p targeting MECP2. Mol. Oncol. 14, 2231–2250. https://doi.org/10.1002/1878-0261.12700 (2020).
Jin, J. et al. miR-19b-3p inhibits breast cancer cell proliferation and reverses saracatinib-resistance by regulating PI3K/Akt pathway. Arch. Biochem. Biophys. 645, 54–60. https://doi.org/10.1016/j.abb.2018.03.015 (2018).
Shi, Q. et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat. Commun. 11, 5513. https://doi.org/10.1038/s41467-020-19349-w (2020).
Won, K. A. & Spruck, C. Triple-negative breast cancer therapy: Current and future perspectives (review). Int. J. Oncol. 57, 1245–1261. https://doi.org/10.3892/ijo.2020.5135 (2020).
Gradishar, W. J. et al. Breast cancer, version 3.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 18, 452–478. https://doi.org/10.6004/jnccn.2020.0016 (2020).
Cui, C. et al. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol. Cancer 19, 58. https://doi.org/10.1186/s12943-020-01180-y (2020).
Barbari, C. et al. Immunotherapies and combination strategies for immuno-oncology. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21145009 (2020).
Keenan, T. E. & Tolaney, S. M. Role of immunotherapy in triple-negative breast cancer. J. Natl. Compr. Cancer Netw. 18, 479–489. https://doi.org/10.6004/jnccn.2020.7554 (2020).
Hegde, P. S. & Chen, D. S. Top 10 challenges in cancer immunotherapy. Immunity 52, 17–35. https://doi.org/10.1016/j.immuni.2019.12.011 (2020).
Braud, V. M. et al. Expression of LLT1 and its receptor CD161 in lung cancer is associated with better clinical outcome. Oncoimmunology 7, e1423184. https://doi.org/10.1080/2162402x.2017.1423184 (2018).
Sawaki, M., Shien, T. & Iwata, H. TNM classification of malignant tumors (Breast Cancer Study Group). Jpn. J. Clin. Oncol. 49, 228–231. https://doi.org/10.1093/jjco/hyy182 (2019).
Giuliano, A. E. et al. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67, 290–303. https://doi.org/10.3322/caac.21393 (2017).
Houston, L., Yu, P., Martin, A. & Probst, Y. Heterogeneity in clinical research data quality monitoring: A national survey. J. Biomed. Inform. 108, 103491. https://doi.org/10.1016/j.jbi.2020.103491 (2020).
Felin, T., Koenderink, J., Krueger, J. I., Noble, D. & Ellis, G. F. R. Data bias. Genome Biol. 22, 59. https://doi.org/10.1186/s13059-021-02278-2 (2021).
Mittelstraß, K. & Waldenberger, M. DNA methylation in human lipid metabolism and related diseases. Curr. Opin. Lipidol. 29, 116–124. https://doi.org/10.1097/mol.0000000000000491 (2018).
Schübeler, D. Function and information content of DNA methylation. Nature 517, 321–326. https://doi.org/10.1038/nature14192 (2015).
Paclik, D. et al. Galectin-2 induces apoptosis of lamina propria T lymphocytes and ameliorates acute and chronic experimental colitis in mice. J. Mol. Med. 86, 1395–1406. https://doi.org/10.1007/s00109-007-0290-2 (2008).
Hoos, A. Development of immuno-oncology drugs—From CTLA4 to PD1 to the next generations. Nat. Rev. Drug Discov. 15, 235–247. https://doi.org/10.1038/nrd.2015.35 (2016).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668. https://doi.org/10.1038/s41577-020-0306-5 (2020).
Miggelbrink, A. M. et al. CD4 T-cell exhaustion: Does it exist and what are its roles in cancer?. Clin. Cancer Res. 27, 5742–5752. https://doi.org/10.1158/1078-0432.Ccr-21-0206 (2021).
Kim, J., Manspeaker, M. P. & Thomas, S. N. Augmenting the synergies of chemotherapy and immunotherapy through drug delivery. Acta biomaterialia 88, 1–14. https://doi.org/10.1016/j.actbio.2019.02.012 (2019).
Bagherifar, R. et al. Nanoparticle-mediated synergistic chemoimmunotherapy for tailoring cancer therapy: Recent advances and perspectives. J. Nanobiotechnol. 19, 110. https://doi.org/10.1186/s12951-021-00861-0 (2021).
Limagne, E. et al. MEK inhibition overcomes chemoimmunotherapy resistance by inducing CXCL10 in cancer cells. Cancer Cell 40, 136-152.e112. https://doi.org/10.1016/j.ccell.2021.12.009 (2022).
O’Donnell, J. S., Teng, M. W. L. & Smyth, M. J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 16, 151–167. https://doi.org/10.1038/s41571-018-0142-8 (2019).
Gote, V., Nookala, A. R., Bolla, P. K. & Pal, D. Drug resistance in metastatic breast cancer: Tumor targeted nanomedicine to the rescue. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22094673 (2021).
Baldominos, P. et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell 185, 1694-1708.e1619. https://doi.org/10.1016/j.cell.2022.03.033 (2022).
Twyman-Saint Victor, C. et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377. https://doi.org/10.1038/nature14292 (2015).
Zhang, Y. & Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 17, 807–821. https://doi.org/10.1038/s41423-020-0488-6 (2020).
Alemohammad, H. et al. The importance of immune checkpoints in immune monitoring: A future paradigm shift in the treatment of cancer. Biomed. Pharmacother. 146, 112516. https://doi.org/10.1016/j.biopha.2021.112516 (2022).
Jiang, Z. H. et al. A pan-cancer analysis reveals the prognostic and immunotherapeutic value of stanniocalcin-2 (STC2). Front. Genet. 13, 927046. https://doi.org/10.3389/fgene.2022.927046 (2022).
Acknowledgements
Thank “XIAN TAO XUE SHU (https://www.xiantao.love/)”, “Home for Researchers editorial team (https://www.home-for-researchers.com)” and “The University of Alabama at Birmingham Cancer data analysis Portal (http://ualcan.path.uab.edu/)”.
Funding
This study is supported by National Key Research and Development (R&D) Program of China (No. 2022YFF0710503, to Y.D.), Jilin Provincial Science and Technology Development Plan Project (No.20230505037ZP, to Y.D.), Jilin Natural Science Fund Project (No. 20210101364JC, to Y.D.), Jilin Provincial Science and Technology Development Plan Project (No. 20210506021ZP, to J.C.).
Author information
Authors and Affiliations
Contributions
R.W. and Y.D. were responsible for the main experimental concept and design. S.H., Z.J., and Q.Z. performed the analysis. S.H., Z.J., Q.Z., and X.Z. prepared the first version of the manuscript. S.H., Z.J., J.C. J.H., and Y.D. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, S., Ji, Z., Zhang, Q. et al. Investigation of LGALS2 expression in the TCGA database reveals its clinical relevance in breast cancer immunotherapy and drug resistance. Sci Rep 13, 17445 (2023). https://doi.org/10.1038/s41598-023-44777-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-44777-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.