Abstract
The biomineral haemozoin, or its synthetic analogue β-haematin (βH), has been the focus of several target-based screens for activity against Plasmodium falciparum parasites. Together with the known βH crystal structure, the availability of this screening data makes the target amenable to both structure-based and ligand-based virtual screening. In this study, molecular docking and machine learning techniques, including Bayesian and support vector machine classifiers, were used in sequence to screen the in silico ChemDiv 300k Representative Compounds library for inhibitors of βH with retained activity against P. falciparum. We commercially obtained and tested a prioritised set of inhibitors and identified the coumarin and iminodipyridinopyrimidine chemotypes as potent in vitro inhibitors of βH and whole cell parasite growth.
Similar content being viewed by others
Introduction
Malaria is an ancient parasitic infection caused by five virulent species of the genus Plasmodium, the most lethal of which is Plasmodium falciparum (Pf)1. Over the past several decades, the emergence of antimalarial-resistant strains of Pf has countered unified efforts to control the disease, and recent reports of partial artemisinin resistance in East Africa have spurred much concern2,3. Although antimalarial research has produced numerous lead candidates, conventional drug discovery strategies such as high-throughput screening (HTS) are notoriously expensive and high risk, and novel drugs have been slow to enter the clinical market4. In silico screening is now a widely used tactic for fast-tracking drug development. With the use of either structure-based or ligand-based methods, large chemical libraries can be virtually screened for compounds most likely to show activity, resulting in significant enrichment rates relative to random screening5.
It is now well-accepted that the 4-aminoquinoline antimalarials, some of which have been rendered ineffective by resistance in certain regions, act by inhibiting haemozoin (HZ) formation6. During the asexual blood stage of the Plasmodium life cycle, haemoglobin is catabolised within the parasite’s digestive vacuole, releasing free haem as a by-product. Haem has the tendency to partition into cell membranes and catalyses the formation of reactive oxygen species; hence, Plasmodium has evolved a complex mechanism of haem detoxification that incorporates the conversion of haem to inert HZ crystals. Given that the combined potency and low cost of chloroquine (CQ), one of the most successful drugs in this class, has yet to be emulated by other clinical antimalarials7, there has been considerable interest in discovering alternative HZ-inhibiting chemotypes. More importantly, compared to other validated targets, HZ has the distinct advantage of not being a gene product, and is thus immutable. Resistance to HZ inhibitors is conferred by mutations in membrane transport proteins that cause an efflux of the drug away from the digestive vacuole, and the structure-specific nature of this mechanism reduces the likelihood of cross-resistance with chemically unrelated HZ inhibitors8. Consequently, a detergent-mediated, biomimetic assay for quantifying β-haematin (synthetic HZ, βH) inhibition was developed and adapted for HTS9,10,11. These screens have been successful in identifying potent βH-inhibiting chemotypes that are active on whole cell Pf cultures, including the benzimidazoles, benzamides and triaryl imidazoles12,13,14,15.
The crystal structure and morphology of βH has also been pertinent in elucidating the mode of action of the 4-aminoquinolines and other βH-inhibiting compounds16. βH comprises centrosymmetrically related haematin ([Fe(III)PPIX]) units which dimerise via two reciprocal iron–carboxylate bonds. In turn, the dimers stack via hydrogen bonds to form parallel strands of porphyrin units17. Buller et al. proposed a non-covalent binding site on the (001) face, which is not only the fastest-growing face and thus the most efficient site for inhibition, but the corrugated surface also exposes chemical groups and aromatic surfaces that favour the adsorption of inhibitors16.
The knowledge of the βH crystal structure and the availability of HTS βH inhibition data makes the target amenable to both structure- and ligand-based virtual screening (S- and LBVS), and in fact, both strategies have previously achieved very promising results. Molecular docking is a structure-based approach that is most commonly used to model the interactions between a small molecule and a protein; however, it can be extended to other classes of therapeutic targets where the structure is known18. A set of drug-like compounds from the ZINC15 database was docked against the βH crystal structure and a small selection of high-ranking compounds was prioritized for experimental testing, resulting in a hit rate of 20% for βH inhibition ≤150 µM19. However, in vitro βH inhibitors are not guaranteed to show activity against whole-cell Pf, and the mechanisms underpinning HZ inhibition are not yet sufficiently understood to rationalise by inspection whether or not a βH inhibitor will be active against the parasite. Furthermore, there are multiple factors that affect the ability of the compound to access and accumulate at the site of the haem target in the DV, so it is uncommon to see direct correlations between βH inhibition activity and whole-cell activity20,21. In this regard, LBVS is an attractive strategy for enriching antiplasmodium hit rates. Typically used in the absence of a 3D target structure, ligand-based methods analyse known small-molecular inhibitors and attempt to correlate their specific structural and physiochemical features with a desired biological activity. Quantitative structure-activity relationships, pharmacophore modelling, and machine learning classifiers are types of ligand-based methods that are commonly used in drug design22. Previously, Bayesian models were built to predict βH-inhibition and antiplasmodium activity and achieved hit rates of 25% and 33%, respectively23.
In this study, we employ a two-step virtual screening workflow involving molecular docking and machine learning methods to screen a commercial library for βH inhibitors with retained activity against cultures of Pf.
Results and discussion
Molecular docking
Molecular docking was used to predict the interaction strength of 25,000 compounds with the in silico βH crystal structure (Fig. 1A). The ChemDiv 300k Representative Compounds library was initially filtered for druglikeness using Lipinski’s Rule of Five and OSIRIS DataWarrior’s24 inhouse score. To further reduce the computational expense of the virtual screen, the “select diverse set” feature in DataWarrior, which uses a fragment-based molecular descriptor to compute structural similarity, was used to obtain a set of 25,000 prioritised compounds. Using Schrodinger’s LigPrep25, the ligands were prepared for docking by performing an energy minimisation and generating their protonated states at pH 5, in accordance with the acidity of the parasite’s digestive vacuole. The ligands were then docked using AutoDock Vina26. A cut-off of − 12 kcal mol−1 was chosen by fitting the docking scores to a cubic function and rounding the inflection point of −12.9 kcal mol−1 up to the nearest whole number (Supplementary Fig. S1A), which classified 1592 molecules (6.4%) as “docking hits” or predicted βH inhibitors. The docking poses show that compounds binding at the (001) face, or the opposite (00\(\stackrel{\mathrm{-}}{1}\)) face have stronger binding affinities, which can be explained by the parallel porphyrin rings and free carboxyl groups that are available for forming π–π stacking interactions and hydrogen bonds, respectively (Supplementary Fig. S1B). Furthermore, the 25,000 compounds were mapped in chemical space using principal component analysis (PCA, Supplementary Fig. S1C), which showed the high-ranking compounds to be negatively shifted in PC1 relative to those with poorer, less negative scores. Based on the PC loadings, this corresponds to molecules with higher logP values and more aromatic rings (Supplementary Table S1), which supports the importance of π–π stacking.
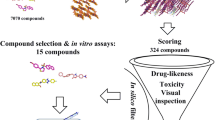
Virtual screening of the ChemDiv 300k Representative Compounds library and prioritisation of in-silico hits for purchasing and experimental testing against βH and whole-cell cultures of Pf. (a) Summarised computational workflow for predicting βH inhibition and antiplasmodium activity. (b) Similarity map of the 374 active compounds from the training set (black) and the 404 LBVS hits, which are categorised as SVM (S, in red), Bayesian (B, in green), and overlap (O, in blue) based on which model(s) predicted them active, overlayed in chemical space. The map assisted in prioritising a set of 24 structurally unrelated compounds which excluded those scaffolds that have been previously investigated as βH inhibitors, i.e., those that appear frequently in the training set actives. (c) Purchased compound S4 docked at (001) face of βH. S4 forms multiple π-π stacking interactions (pink dashed line) with the parallel porphyrin rings and an (amine)NH···O(carboxyl) hydrogen bond (green dashed line).
Building models and in-silico screening for antiplasmodium activity
Two classification models were trained with data from previous high-throughput screens for βH inhibitors (Supplementary Table S2) and used to predict which of the docking hits would be active against whole-cell cultures of Pf. The support vector machine (SVM) model was built using LibSVM, as implemented in the KNIME Analytics Platform27,28. The class-distinguishing ability of four kernel functions (linear, polynomial, radial basis function, sigmoidal) was evaluated by assembling a fivefold cross-validation loop; the training set was partitioned into five groups and for each iteration, one group was held out as the test set such that the activity of each molecule was predicted only once and a receiver operating characteristic (ROC) score for the model was obtained. The linear kernel function showed the best performance, which is not unexpected when modelling a high number of input features N. Here, these features included a total of 1875 1D, 2D, and 3D molecular descriptors and the extended connectivity fingerprint (ECFP6). The regularization parameter C, which guards against over-fitting by controlling the model’s tolerance for misclassified data, was optimised to a value of 0.274, which resulted in a ROC score of 0.935 (Supplementary Table S3). For the Bayesian Fingerprint model, the performance of various molecular fingerprints was evaluated, with the circular functional connectivity (FCFP6) descriptor achieving the best ROC score of 0.918 (Supplementary Table S4). Due to the reduced computational expense of building Bayesian models relative to the SVM, here the model was optimised via a leave-one-out cross-validation loop in which molecules were individually held out as the test set.
The optimised SVM and Bayesian models were then used screen the 1592 docking hits for antiplasmodium activity, which predicted a total of 404 of these (25%) to be bioactive. These were categorised as: SVM and Bayesian overlap (O), Bayesian only (B), or SVM only (S) based on which models predicted them to be active. Interestingly, when the models were used to predict the activity of the control compound, chloroquine (Supplementary Table S5), the SVM model falsely classified the drug as inactive. This could be explained by chloroquine’s long, flexible side chain at position 4 of the quinoline ring, which distinguishes it from the highly lipophilic, planar βH inhibitors that dominate the active class in the training set. In contrast, the Bayesian Fingerprint model correctly predicted chloroquine to be active.
Once the models had been optimised, the predictions made for the training set were analysed to identify substructures that appear frequently in each of the respective classes (Table 1). The Bayesian Fingerprint and SVM models detect the same fragments as being favourable for antiplasmodium activity, several of which comprise an N-heteroaromatic ring. The urea moiety is also significantly enriched in the active class. Conversely, the 1,4-dihydroquinoline scaffold and several of its derivatives frequently appear in the inactive class, as do the aryl sulfonamides.
The 404 in-silico hits comprise a total of 73 scaffolds, 41 of which are not present in the training set. However, 13 of these unique scaffolds have previously documented antiplasmodium activity (Fig. 2 and Supplementary Fig. 2), including the pyrido[1,3-d]pyrimidin-4-one and 1,2,4-triazino[5,6-b]indole scaffolds that were previously identified as haemozoin inhibitors. Other targets which have been proposed for particular scaffolds (Fig. 2) include deoxyhypusine hydroxylase, glucose-6-phosphate dehydrogenase, dihydrofolate reductase, cysteine protease, and other elements of the haem detoxification pathway, while the modes of the action for the remaining scaffolds have not yet been elucidated (Supplementary Fig. 2)29,30,31,32,33,34,35.
Similarity analysis, visual inspection of docking poses, and compound selection
All 404 compounds could not be purchased, so further criteria were introduced to prioritise compounds for experimental testing. Firstly, the compounds were ranked by their SVM and Bayesian scores and the top compounds were examined. This set had limited structural diversity and was notably dominated by chemotypes with well-documented βH inhibition and antiplasmodium activity, i.e., quinolines and benzimidazoles. Thus, a similarity map of both the LBVS hits and the actives –from the training set was generated in OSIRIS DataWarrior24 (Fig. 1B) to assist in the selection of a structurally diverse, representative set of compounds that was enriched in novel chemotypes. Compounds of the same chemical class were distinguished by their in-silico scores, as well as their docking poses, which were inspected in Discovery Studio Visualizer36. Preference was given to compounds showing multiple π–π stacking interactions with the porphyrin rings of the crystal and/or hydrogen bonds with the free carboxyl side chains, as well as those docking to the crystal via two haematin units, which is facilitated by a twist between the two aromatic moieties in the molecule (Fig. 1C). A total of 31 compounds were purchased, which included eight compounds from each of the O and B categories, seven from the S, and eight classified as “non-hits”. Four of these were docking hits that were predicted inactive by both antiplasmodium models, and the remaining docked with poor scores between –4.5 and –6 kcal mol−1 to the in silico βH crystal structure (Fig. 3 and Supplementary Table S5).
NP-40 detergent-mediated assay for βH inhibition
The compounds were tested for activity against βH at a concentration range of 0–500 µM, using the detergent-mediated and colorimetric pyridine ferrochrome methods described by Carter et al. and Ncokazi and Egan, respectively (Supplementary Table S6)9,10. As expected, the four compounds that received poor docking scores (−4.5 to −6 kcal mol−1) showed no activity against βH up to a concentration of 500 µM. Of the 27 compounds classified as docking hits, 11 showed IC50 values < 150 µM, corresponding to a hit rate of 41% for βH inhibition. Five compounds showed IC50 values comparable to CQ (16-40 μM) and O3 and I6 showed superior activity with IC50s of 8 and 11 μM, respectively. Nine of the eleven βH inhibitors were classified as LBVS hits (Table 2).
pLDH assay for Pf growth inhibition
All 31 purchased compounds were tested at concentrations of 1 and 5 µM against the CQ-sensitive Pf NF45 strain, using the parasite lactate dehydrogenase (pLDH) assay described by Makler et al (Supplementary Table S7 and S8)35,37. Interestingly, one compound predicted to be inactive against parasites by both the SVM and Bayesian models showed moderate growth inhibition (∼ 60%) at both tested concentrations. This compound was not a βH inhibitor, so it likely acts against other biological targets or via a different pathway. Since the antiplasmodium models were trained only on βH inhibiting active compounds, this would explain why neither model predicted this compound to be active. Of the nine βH inhibitors, eight inhibited Pf growth >50% at 5 µM. Excluding O5, the activity of these compounds was fully ascertained by performing dose-response assays against the Pf NF54 and CQ-resistant Pf Dd2 strains (Supplementary Table S8). Six compounds showed IC50 values ≤ 5 against both strains, corresponding to an overall hit rate of 30% for antiplasmodium activity. Furthermore, compound B1 showed sub-micromolar activity against the Dd2 strain and compounds O3 and S1 showed sub-micromolar activity against both the NF54 and Dd2 strains (Table 2). The primary mode of action for these compounds is presumed to be via the inhibition of hemozoin formation and a subsequent increase in cytotoxic haem or drug-haem complexes, however the disruption of other biological pathways may contribute to their activity. This would need further investigation on a case-by-case basis to validate the biological mechanisms or involvement of protein targets for each compound.
Interestingly, compounds O3 and O8 both contain a coumarin ring system. The structural diversity and abundance of coumarins isolated from natural sources has drawn particular interest from the pharmaceutical industry. The coumarin core is considered a privileged scaffold, particularly since these compounds have displayed antimicrobial, anticancer, and antioxidant activities. In addition to some coumarin-containing plant extracts, synthetic coumarin derivatives have been investigated as potential antimalarial agents, with success mostly being found via molecular hybridization. For example, chalcone-triazole- and ferrocenyl-oxazine-coumarins were found to inhibit Pf growth in the low micromolar range. The respective authors cite falcipain-2 inhibition and DNA binding as potential targets, with haemozoin inhibition also implicated for the latter series38,39. The training set contains a total of fifteen coumarins, which all showed βH IC50 values < 60 μM, but are inactive against whole-cell Pf cultures. It is interesting that, despite the absence of this chemotype in the “active” training set, both antiplasmodium models identified two experimentally active coumarin-containing molecules. Further, of the 81 coumarins classified as docking hits, an additional 17 were predicted active by the SVM and/or Bayesian models.
Compound S1 contains the iminodipyridinopyrimidine (IDPP) core and is particularly interesting in that there has been very little investigation into the antimalarial potential of this scaffold. However, S1 is reported as antimalarially active in the PubChem BioAssay database and was found to inhibit parasite growth in the first generation (48 h incubation) with an EC50 of 0.5 µM (AID 504832). Several analogues of these compounds are also contained in the BioAssay database and have exhibited good to moderate activities against Pf 3D7 and Dd2 (AID 2302 & 2306), although the biological target and structure-activity relationships of this compound class have yet to be investigated. The IDPP scaffold was not present in the training set. Twenty-nine IDPPs were present in the set of 25 000 compounds screened, with seven having docking scores ≤ –12 kcal/mol. Of these, only S1 and its analogue were predicted bioactive by either classification model. These differ from the remaining five IDPPs in the presence of a basic nitrogen on the substituent of the dihydropyridyl ring, which is predicted to be ~ 95% protonated at pH 5, and only 15–18% protonated at pH 7. Chloroquine’s potency is often partially attributed to ‘pH trapping’, the mechanism by which the drug accumulates in the parasite’s digestive vacuole. It is possible that the SVM model implicitly characterised a molecule’s propensity for pH trapping as being important for antiplasmodium activity.
Conclusions
Modern drug discovery is becoming increasingly in silico based to mitigate the exorbitant costs of conventional HTS. This study capitalises on the previously solved βH crystal structure and the abundance of publicly available HTS data by combining SB- and LBVS techniques to identify βH inhibitors with retained activity against Pf. This two-step workflow achieved excellent enrichment rates for βH inhibition relative to random screening. The antiplasmodium SVM model in particular was successful in identifying three compounds with sub-micromolar activity against Pf. The coumarin and IDPP scaffolds represent promising starting points for lead optimisation and merit further pharmacological investigation.
Computational and experimental methods
Molecular docking for potential β-haematin inhibitors
Preparing the commercial library
Hierarchical virtual screening was carried out on the ChemDiv 300k Representative Compounds Library40; first by filtering for druglikeness in OSIRIS DataWarrior24. Lipinski’s Rule of Five, a widely used estimator for oral bioavailability, was applied to exclude molecules in violation of any the following criteria: molecular weight ≤ 500 Da, number of hydrogen bond donors ≤ 5, number of hydrogen bond acceptors ≤ 10, octanol-water partition coefficient (logP) ≤ 5. DataWarrior’s in-house ‘druglikeness’ score was used as an additional filter, excluding molecules that received < 0. Finally, the ‘select diverse set’ feature was implemented to obtain a small representative library of 25,000 molecules.
Preparing ligands for molecular docking
The filtered library was prepared for molecular docking in Maestro, an interface for the Schrödinger computational platform41. LigPrep was used to convert the 2D structures to their energy minimised 3D conformers with the OPLS3e force field25. Using Epik, the protonated species were generated at pH 5.0 ± 0.5, based on the pH of the biological target of interest i.e., the Pf digestive vacuole42.
The receptor model for β-haematin
Using a modified cvff force field, the βH µ-propianato dimer was optimised by a group of researchers at Stellenbosch University with the BIOVIA Materials Studio package43,44. A receptor model expressing the dominant (100), (010) and fastest-growing (001) faces was generated, using the same software, by growing a 3 × 3 × 3 ‘supercell’ and exporting it as a Protein Data Bank (pdb) file.
Molecular docking against the β-haematin crystal structure
Docking was performed in the Python Prescription Virtual Screening Tool (PyRx), which compiles several open-source programs into one user-friendly interface, including OpenBabel and AutoDock Vina26. As required by AutoDock Vina, all receptor and ligand structures were converted to pdbqt format, an extension of the pdb format with partial charges (Q) and atom types (T) defined. Within the Vina wizard, the search space was set to enclose the entirety of the crystal surface; centre (x, y, z): (13.5, 22.5, 12) and dimensions (x, y, z): (48, 47.5, 43). Each ligand was docked at an exhaustiveness of 8 and only the lowest energy binding mode was retained. Upon visual inspection of the docking scores, a Vina binding affinity of −12 kcal mol−1 was chosen as the cut-off for hit selection, classifying 1592 compounds as potential βH inhibitors.
Classification models for predicting antiplasmodium activity
The antiplasmodium models were built in the KNIME Analytics Platform v4.3.327. Though not developed specifically for drug discovery, KNIME’s collaborative philosophy has meant that several cheminformatics platforms, including RDKit and the Chemistry Development Kit (CDK), have integrations within the platform. Together with its drag-and-drop style graphical interface, this makes KNIME an attractive machine learning tool for non-experts.
Training data
The training data was largely sourced from a HTS for βH-inhibiting antimalarials, piloted by Vanderbilt University (VU)15. Only βH inhibitors (βH IC50 ≤ 100 µM) which had been tested against Pf were included; molecules were considered ‘active’ (1) if they exhibited Pf IC50 ≤ 1 µM and ‘inactive’ (0) if Pf IC50 ≥ 1.5 µM. The same criteria were applied to ∼100 bioactive neo- and isocryptolepine derivatives synthesised by a group at Okayama University (OU)45,46. In addition, a number of molecules from the Tres Cantos Antimalarial Compounds Set (TCAMS)47 were included in the training set using unpublished βH inhibition data; these molecules all exhibited ≥ 90% βH inhibition and were considered active if they inhibited ≥ 90% Pf growth, both relative to the chloroquine control drug. The resultant set contained a total of 1606 molecules, with 374 actives (Supplementary Table S2). For the SVM model, the training molecules were converted to their 3D energy minimised representations at pH 5 ± 0.5 in LigPrep and multiple protonation states were retained, resulting in 1707 data instances.
Molecular descriptors
Five types of molecular fingerprints were generated with the CDK’s Fingerprints node in KNIME. In addition, a total of 1875 molecular descriptors (1D, 2D and 3D) were calculated using the open-source software PaDEL.
Support vector machine (SVM)
The C-SVM classifier was built using LibSVM, an open-source library that has its learning code implemented in KNIME28. The input features were prepared by (a) expanding the ECFP6 fingerprint into a series of 1024 integers and (b) normalising the 1875 descriptors calculated in PaDEL. Since there is no sure method for predicting which kernel function will perform best for a given dataset, each kernel implemented in LibSVM (linear, polynomial, RBF, sigmoidal) was evaluated via a fivefold cross-validation loop with stratified sampling. The relevant hyperparameters were optimised for each kernel via the hillclimbing method by incorporating a parameter optimisation loop.
Bayesian classification model
The Bayesian classifier was built using the Fingerprint Bayesian Learner and Predictor nodes. Generally, Bayesian models utilise a naïve, Laplacian-corrected algorithm based on Bayes theorem of conditional probability.
where P(A|B) is the probability of a compound being active given the presence of a molecular feature, P(B|A) is the likelihood of a feature being present in an active compound, P(A) is the probability of a compound in the training set being active, and P(B) is the probability of a feature being present in the training set. The classifier ‘naively’ assumes that the input features are independent and multiplies the probabilities of the individual events. However, the frequency of features in the training set is accounted for by introducing a Laplacian-corrected estimator, so that the score is given as a sum of the corrected estimators. Models were built for five molecular fingerprints implemented in the CDK and evaluated by a leave-one-out cross validation loop in which molecules are individually held out and classified using the remaining data instances.
NP-40 detergent mediated assay for βH formation
A set of 31 compounds was purchased based on their in-silico scores, structural diversity, and availability. The βH inhibition activity of the purchased compounds was investigated using the detergent-mediated NP-40 assay developed by Carter et al. in 96-well plates10. The assay was analysed using the pyridine-ferrochrome method described by Ncokazi and Egan9. The UV-vis absorbance was read at 405 nm on a Thermo Scientific Multiskan GO plate reader and the IC50 values were calculated by plotting sigmoidal dose-response curves in GraphPad Prism v 9.0.0. (GraphPad Software Inc., La Jolla, CA, USA).
Parasite lactate dehydrogenase assay for antiplasmodium activity
The growth inhibition activity of the compounds was tested against two Pf strains: CQ-sensitive NF54 and CQ-resistant Dd2 cell lines. Dose response activity was measured with the pLDH assay in 96-well plates, as described by Makler et al. in 96-well plates37. The UV-vis absorbance was read at 620 nm on a MultiSkan Go plate reader and IC50 values were determined using non-linear dose-response analysis in GraphPad Prism v 9.0.0.
Data availability
The datasets used and/or analysed during the current study are available in the supplementary information files or from the corresponding author on reasonable request.
References
Snow, R. W. Global malaria eradication and the importance of Plasmodium falciparum epidemiology in Africa. BMC Med. 13, 23. https://doi.org/10.1186/s12916-014-0254-7 (2015).
Blasco, B., Leroy, D. & Fidock, D. A. Antimalarial drug resistance: Linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 23, 917–928. https://doi.org/10.1038/nm.4381 (2017).
Stokes, B. H. et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. eLife 10, e66277. https://doi.org/10.7554/eLife.66277 (2021).
Wells, T. N. C., van Huijsduijnen, R. H. & Van Voorhis, W. C. Malaria medicines: A glass half full?. Nat. Rev. Drug Discov. 14, 424–442. https://doi.org/10.1038/nrd4573 (2015).
Yan, X. C. et al. Augmenting hit identification by virtual screening techniques in small molecule drug discovery. J. Chem. Inf. Model. 60, 4144–4152. https://doi.org/10.1021/acs.jcim.0c00113 (2020).
Herraiz, T., Guillén, H., González-Peña, D. & Arán, V. J. Antimalarial quinoline drugs inhibit β-hematin and increase free hemin catalyzing peroxidative reactions and inhibition of cysteine proteases. Sci. Rep. 9, 15398. https://doi.org/10.1038/s41598-019-51604-z (2019).
Medicine, I. O. Saving Lives, Buying Time: Economics of Malaria Drugs in an Age of Resistance (The National Academies Press, 2004).
Ecker, A., Lehane, A. M., Clain, J. & Fidock, D. A. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 28, 504–514. https://doi.org/10.1016/j.pt.2012.08.002 (2012).
Ncokazi, K. & Egan, T. A colorimetric high-throughput β-hematin inhibition screening assay for use in the search for antimalarial compounds. Anal. Biochem. 338, 306–319. https://doi.org/10.1016/j.ab.2004.11.022 (2005).
Carter, M. D., Phelan, V. V., Sandlin, R. D., Bachmann, B. O. & Wright, D. W. Lipophilic mediated assays for beta-hematin inhibitors. Comb. Chem. High Throughput Screen 13, 285–292. https://doi.org/10.2174/138620710790980496 (2010).
Sandlin, R. D. et al. Use of the NP-40 detergent-mediated assay in discovery of inhibitors of beta-hematin crystallization. Antimicrob. Agents Chemother. 55, 3363–3369. https://doi.org/10.1128/aac.00121-11 (2011).
L’Abbate, F. P. et al. Hemozoin inhibiting 2-phenylbenzimidazoles active against malaria parasites. Eur. J. Med. Chem. 159, 243–254. https://doi.org/10.1016/j.ejmech.2018.09.060 (2018).
Wicht, K. J., Combrinck, J. M., Smith, P. J., Hunter, R. & Egan, T. J. Identification and mechanistic evaluation of hemozoin-inhibiting triarylimidazoles active against Plasmodium falciparum. ACS Med. Chem. Lett. 8, 201–205. https://doi.org/10.1021/acsmedchemlett.6b00416 (2017).
Wicht, K. J., Combrinck, J. M., Smith, P. J., Hunter, R. & Egan, T. J. Identification and SAR evaluation of hemozoin-inhibiting benzamides active against Plasmodium falciparum. J. Med. Chem. 59, 6512–6530. https://doi.org/10.1021/acs.jmedchem.6b00719 (2016).
Sandlin, R. D. et al. Identification of β-hematin inhibitors in a high-throughput screening effort reveals scaffolds with in vitro antimalarial activity. Int. J. Parasitol. Drugs Drug Resist. 4, 316–325. https://doi.org/10.1016/j.ijpddr.2014.08.002 (2014).
Buller, R., Peterson, M. L., Almarsson, Ö. & Leiserowitz, L. Quinoline binding site on malaria pigment crystal: A rational pathway for antimalaria drug design. Cryst. Growth Des. 2, 553–562. https://doi.org/10.1021/cg025550i (2002).
Pagola, S., Stephens, P. W., Bohle, D. S., Kosar, A. D. & Madsen, S. K. The structure of malaria pigment beta-haematin. Nature 404, 307–310. https://doi.org/10.1038/35005132 (2000).
Meng, X. Y., Zhang, H. X., Mezei, M. & Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 7, 146–157. https://doi.org/10.2174/157340911795677602 (2011).
de Sousa, A. C. C., Combrinck, J. M., Maepa, K. & Egan, T. J. Virtual screening as a tool to discover new β-haematin inhibitors with activity against malaria parasites. Sci. Rep. 10, 3374. https://doi.org/10.1038/s41598-020-60221-0 (2020).
Tam, D. N. H. et al. Correlation between anti-malarial and anti-haemozoin activities of anti-malarial compounds. Malar. J. 19, 298. https://doi.org/10.1186/s12936-020-03370-x (2020).
Kaschula, C. H. et al. Structure−activity relationships in 4-aminoquinoline antiplasmodials. The role of the group at the 7-position. J. Med. Chem. 45, 3531–3539. https://doi.org/10.1021/jm020858u (2002).
Acharya, C., Coop, A., Polli, J. E. & Mackerell, A. D. Jr. Recent advances in ligand-based drug design: Relevance and utility of the conformationally sampled pharmacophore approach. Curr. Comput. Aided Drug Des. 7, 10–22. https://doi.org/10.2174/157340911793743547 (2011).
Wicht, K. J., Combrinck, J. M., Smith, P. J. & Egan, T. J. Bayesian models trained with HTS data for predicting β-haematin inhibition and in vitro antimalarial activity. Bioorg. Med. Chem. 23, 5210–5217. https://doi.org/10.1016/j.bmc.2014.12.020 (2015).
Sander, T., Freyss, J., von Korff, M. & Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 55, 460–473. https://doi.org/10.1021/ci500588j (2015).
Schrödinger Release 2022-1. LigPrep. (Schrödinger, LLC, 2021).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. https://doi.org/10.1002/jcc.21334 (2010).
Berthold, M. R. et al. Data Analysis, Machine Learning and Applications. (eds. Preisach, C., Burkhardt, H., Schmidt-Thieme, L. & Decker, R). 319–326 (Springer).
Chang, C.-C. & Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 27. https://doi.org/10.1145/1961189.1961199 (2011).
Kgokong, J. L., Smith, P. P. & Matsabisa, G. M. 1,2,4-Triazino-[5,6b]indole derivatives: Effects of the trifluoromethyl group on in vitro antimalarial activity. Bioorg. Med. Chem. 13, 2935–2942. https://doi.org/10.1016/j.bmc.2005.02.017 (2005).
Miguel-Blanco, C. et al. The antimalarial efficacy and mechanism of resistance of the novel chemotype DDD01034957. Sci. Rep. 11, 1888. https://doi.org/10.1038/s41598-021-81343-z (2021).
Preuss, J. et al. Discovery of a Plasmodium falciparum glucose-6-phosphate dehydrogenase 6-phosphogluconolactonase inhibitor (R, Z)-N-((1-ethylpyrrolidin-2-yl)methyl)-2-(2-fluorobenzylidene)-3-oxo-3,4-dihydro-2H-benzo[b][1,4]thiazine-6-carboxamide (ML276) that reduces parasite growth in vitro. J. Med. Chem. 55, 7262–7272. https://doi.org/10.1021/jm300833h (2012).
Hicks, R. et al. Evaluation of 4-azaindolo[2,1-b]quinazoline-6,12-diones’ interaction with hemin and hemozoin: A spectroscopic, X-ray crystallographic and molecular modeling study. Internet Electron. J. Mol. Des. 4, 751–764 (2005).
Gujjar, R. et al. Lead optimization of aryl and aralkyl amine-based triazolopyrimidine inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase with antimalarial activity in mice. J. Med. Chem. 54, 3935–3949. https://doi.org/10.1021/jm200265b (2011).
Thakur, R. K. et al. Synthesis and antiplasmodial activity of glyco-conjugate hybrids of phenylhydrazono-indolinones and glycosylated 1,2,3-triazolyl-methyl-indoline-2,3-diones. Eur. J. Med. Chem. 155, 764–771. https://doi.org/10.1016/j.ejmech.2018.06.042 (2018).
Charris, J. et al. Synthesis and evaluation as antimalarials of thieno-[2,3-b]quinoline 2-carboxate acid derivatives. Ciencia 15, 64 (2007).
BIOVIA, D.S. Discovery Studio Visualizer, v21. 1.0., 2020. (Dassault Systmès BIOVIA, 2010).
Makler, M. T. et al. Parasite lactate dehydrogenase as an assay for Plasmodium falciparum drug sensitivity. Am. J. Trop. Med. Hyg. 48, 739–741. https://doi.org/10.4269/ajtmh.1993.48.739 (1993).
Mbaba, M. et al. Coumarin-annulated ferrocenyl 1,3-oxazine derivatives possessing in vitro antimalarial and antitrypanosomal potency. Molecules. https://doi.org/10.3390/molecules26051333 (2021).
Bouckaert, C. et al. Synthesis, evaluation and structure–activity relationship of new 3-carboxamide coumarins as FXIIa inhibitors. Eur. J. Med. Chem. 110, 181–194. https://doi.org/10.1016/j.ejmech.2016.01.023 (2016).
ChemDiv Inc. http://www.chemdiv.com/. (ChemDiv Inc.).
Schrödinger Release 2022-1. Maestro (Schrödinger, LLC, 2021).
Schrödinger Release 2022-1. Epik. (Schrödinger, LLC, 2021).
BIOVIA, D.S. Materials Studio, 6.0. (Dassault Systmès BIOVIA).
Olivier, T. et al. Adsorption to the surface of hemozoin crystals: Structure-based design and synthesis of new amino-phenoxazine β-hematin inhibitors. ChemMedChem https://doi.org/10.1002/cmdc.202200139 (2022).
Wang, N. et al. Synthesis and in vitro testing of antimalarial activity of non-natural-type neocryptolepines: Structure–activity relationship study of 2,11- and 9,11-disubstituted 6-methylindolo[2,3-b]quinolines. Chem. Pharm. Bull. (Tokyo) 61, 1282–1290. https://doi.org/10.1248/cpb.c13-00639 (2013).
Wang, N. et al. Synthesis, β-haematin inhibition, and in vitro antimalarial testing of isocryptolepine analogues: SAR study of indolo[3,2-c]quinolines with various substituents at C2, C6, and N11. Bioorg. Med. Chem. 22, 2629–2642. https://doi.org/10.1016/j.bmc.2014.03.030 (2014).
Gamo, F. J. et al. Thousands of chemical starting points for antimalarial lead identification. Nature 465, 305–310. https://doi.org/10.1038/nature09107 (2010).
Acknowledgements
This research was funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number 5R01AI143521. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also funded by the National Research Foundation and the Harry Crossley Foundation. We thank the Centre for High Performance Computing (CHPC) for providing access to the Schrödinger computational platform.
Author information
Authors and Affiliations
Contributions
L.A. performed the in-silico screening and NP-40 assay for βH formation, R.M. and N.T. performed the pLDH assay for parasite growth, and T.J.E. and K.W. conceived and directed the study. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amod, L., Mohunlal, R., Teixeira, N. et al. Identifying inhibitors of β-haematin formation with activity against chloroquine-resistant Plasmodium falciparum malaria parasites via virtual screening approaches. Sci Rep 13, 2648 (2023). https://doi.org/10.1038/s41598-023-29273-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29273-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.