Abstract
Ammonia-oxidizing archaea and bacteria (AOA and AOB, respectively) are important intermediate links in the nitrogen cycle. Apart from the AOA and AOB communities in soil, we further investigated co-occurrence patterns and microbial assembly processes subjected to inorganic and organic fertilizer treatments for over 35 years. The amoA copy numbers and AOA and AOB communities were found to be similar for the CK and organic fertilizer treatments. Inorganic fertilizers decreased the AOA gene copy numbers by 0.75–0.93-fold and increased the AOB gene copy numbers by 1.89–3.32-fold compared to those of the CK treatment. The inorganic fertilizer increased Nitrososphaera and Nitrosospira. The predominant bacteria in organic fertilizer was Nitrosomonadales. Furthermore, the inorganic fertilizer increased the complexity of the co-occurrence pattern of AOA and decreased the complexity pattern of AOB comparing with organic fertilizer. Different fertilizer had an insignificant effect on the microbial assembly process of AOA. However, great difference exists in the AOB community assembly process: deterministic process dominated in organic fertilizer treatment and stochastic processes dominated in inorganic fertilizer treatment, respectively. Redundancy analysis indicated that the soil pH, NO3−N, and available phosphorus contents were the main factors affecting the changes in the AOA and AOB communities. Overall, this findings expanded our knowledge concerning AOA and AOB, and ammonia‐oxidizing microorganisms were more disturbed by inorganic fertilizers than organic fertilizers.
Similar content being viewed by others
Introduction
Nitrogen is absorbed and utilized by microorganisms and crops that plays an essential role in regulating the global N cycle. Excessive application of nitrogen fertilizer has deeply changed soil N cycle and as one major factor resulted nitrogen loss. Ammonia oxidation, the key process of the soil N cycle, is chiefly carried out by ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA) and complete ammonia oxidizing bacteria1,2,3. This further affected nitrogen leaching and retention with concomitant impacts on both crop productivity and the environment. Reducing inorganic fertilizer application and increasing organic fertilizer could alleviate the accumulation of soil nitrate nitrogen and improve the nitrogen utilization rate. Research on the microbiome of ammonia‐oxidizing microorganisms on different fertilizer application, as well as interaction mechanism, can help us comprehensively understand soil N cycling and dynamics, and formulate reasonable farmland fertilization strategies, which was of great importance in the reduce of chemical inputs and nitrogen loss.
Their abundance and distribution have been substantially researched in various habitats. However, little is known about the interaction mechanism of AOA and AOB corresponding to long-term fertilization. Studies have shown that excessive use of chemical fertilizers deleteriously changes the soil microbial community4,5. Intensive inorganic fertilizer inputs lead to a shift from fungal-dominated to bacterial soil food webs6, indicating that the capacity of material transformation was reduced7,8. The studies in our lab showed that soil bacterial diversity and abundance decreased due to the application of inorganic fertilization9,10. The AOA and AOB in black soil responded differently to environmental disturbances and resource utilization11,12. A 50-year field trial showed that the type and concentration of organic matter affected the structures of AOB and AOA communities in the clay loam soil13. Shi et al.14 demonstrated that organic fertilizers have significant effects on the diversity and community structure of AOB. All these findings indicate that inorganic and organic fertilizers have different properties, leading to questions about what aspects of AOA and AOB in the soil are influenced by the two fertilizer types and to what extent the AOA and AOB communities can be changed after the application of fertilizers.
In recent years, network analysis has become an accepted advanced research method to analyze the competition or mutually beneficial cooperation between many microbial communities, such as bacteria, fungi, actinomycetes and archaea15,16. The utility of network analysis accounts for potential biotic interactions between soil microorganisms that define the niche space of AOA and AOB17. The majority of nitrite-oxidizing bacteria and AOA formed two independent modules. Hence, changes in ammonia-oxidizing archaeal and bacterial co-occurrence patterns in soil undergoing long-term inorganic and organic fertilizer treatments needs to be investigated. The hub of AOA and AOB could provide further knowledge to understand the key connected microbe in the nitrification process and its mechanism in the northeast black soil.
The microbial assembly process, one of the advanced and prevalent approaches currently adopted in ecology for exploring microbial mechanisms, has obtained extensive research results in recent years18,19,20. The niche-based and neutral theories were adopted to perform quantitative analysis and examine the contributions of deterministic and stochastic processes in the microbial community assembly. While the niche-based theory is based on the differences in ecological niches of co-occurring species21, the neutral theory relies on dispersal and stochastic demographic processes22. Deterministic and stochastic processes simultaneously influence community assembly in various ecosystem types23,24,25. A recent study demonstrated that the assembly processes of AOA and AOB were different, and the balance between these two communities determined the species co-existence in the forest and meadow soils from temperate to tropical regions26. The AOA assembly processes were not altered by the conversion of grassland to cropland; however, the AOB community assembly process shifted from stochastic to deterministic processes in grassland and cropland, respectively27. This study has important implications for the potential diversification of soil functions under environmental changes. Therefore, assessing the long-term fertilizer application and how to differ the assembly process of ammonia-oxidizing microorganisms might provide a new strategy to control the nitrogen leaching and groundwater pollution in farmland.
The objective of this research was to comprehensively evaluate the interaction mechanism among ammonia-oxidizing microorganisms as a result of the introduction of inorganic and organic fertilizers in the black soil. We used quantitative polymerase chain reaction (qPCR) and high-throughput sequencing to detect the changes in AOA and AOB communities. Network analysis and microbial assembly process were used to assess the interactions among AOA and AOB affected by the inorganic and organic fertilizer. This study offers novel insights into the relationship between inorganic and organic fertilizers and ammonia-oxidizing microorganism.
Results
Soybean yield and soil properties
Fertilization improved soybean yield significantly, and the N1, N2 and M treatment increased the yield by 23.58%, 53.37%, and 39.55%, respectively, comparing with CK treatment (Fig. S1). Furthermore, there was no significant difference in yield among the organic (M) and the two inorganic treatments (N1 and N2), indicating that with an abundant nitrogen input, the organic fertilizer could support an ideal yield if used in place of the inorganic fertilizers.
The soil properties of the four treatments were different (Table 1). The pH of CK and M treatment was 6.48 and 6.59, and when the inorganic fertilizer was applied, the pH values of the N1 and N2 treated soils were reduced to 5.47 and 4.62, respectively. This finding indicated that the organic fertilizer could effectively maintain soil pH, whereas the inorganic fertilizer caused a pH reduction. There was more available nitrogen (NO3−-N and NH4+-N) in the fertilizer treatments than in the CK treatment, especially the NO3−-N content, which was higher by 115.90%, 138.16%, and 88.27% in the N1, N2, and M treatments, respectively, compared to that in the CK treatment. The N2 treatment had the highest AP content (64.85 mg kg–1), and the M treatment had the highest AK content (190.20 mg kg–1). The OM contents were higher in all fertilizer treatments than in the CK treatment.
Copy numbers of amoA gene
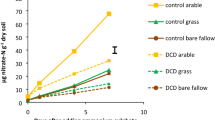
Differences in the AOA and AOB amoA copy numbers among the treatments indicated that the sizes of the communities of ammonia-oxidizing microorganisms was influenced by long-term fertilization differently. In general, the AOA amoA copy numbers in CK and M treatment were much higher than that in N1 and N2; the AOB amoA copy numbers in CK and M treatment were much lower than that in N1 and N2 (Fig. S2). The copy numbers of AOA amoA gene were significantly lower by 75.15% in the N1 (7.99 × 106 copies/g soil) and 93.33% in the N2 (2.15 × 104 copies per ng DNA) treatments compared to those in the CK treatment. In contrast, the AOB amoA copy numbers in the N1 (1.86 × 104 copies/g soil) and N2 (2.78 × 106 copies/g soil) treatments were significantly (1.89 and 3.32-fold, respectively) higher than those in the CK treatment. The AOA and AOB amoA gene copy numbers in the M treatment (3.88 × 106 and 5.37 × 105 copies/g soil, respectively) were slightly lower, but not significantly different from the CK treatment gene copy numbers (AOA: 3.22 × 106 and AOB: 6.44 × 105 copies per ng DNA).
Correlation analysis among AOA and AOB sequence copy numbers and soil properties provided further insight into the factors driving the differences in AOA and AOB amoA copy numbers (Table 2). The AOA amoA copy numbers were negatively correlated with soil NO3−-N (r = − 0.717, p < 0.01) and AP (r = − 0.712, p < 0.01) levels and positively correlated with soil pH (r = 0.893, p < 0.01). However, the AOB amoA copy numbers were positively correlated with soil NO3−-N (r = 0.660, p < 0.05) and AP (r = 0.746, p < 0.01) concentrations and negatively correlated with soil pH (r = − 0.868, p < 0.01). Therefore, soil pH, NO3−-N content, and AP content played vital roles in the changes in amoA copy numbers among all fertilizer treatments.
Alpha diversity of soil AOA and AOB communities
The coverage, diversity and richness indices of the four fertilization treatments are presented in Table S1. The values of the community Chao1 and ACE indices were higher in the fertilizer treatments than those in the CK treatment for both AOA and AOB diversity. Moreover, the fertilizer treatments significantly affected the AOA diversity. The Chao1 index for M, N1 and N2 treatments increased by 15.16% to 19.99% for AOA diversity and by 2.42% to 9.51% for AOB diversity compared to that for the CK treatment. Furthermore, the AOA and AOB Shannon indices were higher for the fertilizer treatments than those for the CK treatment, except for the AOB Shannon index for the M treatment.
The relationships between the AOA and AOB alpha diversity and the soil properties are shown in Table 2. For the AOA, the Shannon indices were negatively correlated with soil TN content (r = − 0.766, p < 0.01), and the Simpson indices were positively correlated with soil NO3−N (r = − 0.643, p < 0.05) and TN (r = 0.666, p < 0.05) contents. In contrast, for the AOB, the Shannon indices were negatively correlated with soil pH (r = − 0.934, p < 0.01) and positively correlated with soil AP levels (r = 0.584, p < 0.05), and the Simpson indices were positively correlated with soil pH (r = 0.957, p < 0.01). These results indicated that alpha diversity of AOA was more sensitive to soil NO3−-N content, whereas alpha diversity of AOB was more sensitive to soil pH.
Beta diversity of the AOA and AOB communities
Analysis revealed that the types of microbes observed were similar across the treatments, but the microbial abundances differed (Fig. S3). The predominant phyla in the AOA communities in the four treatments included Thaumarchaeota, Archaea_unclassified, Crenarchaeota, and environmental_samples. Environmental_samples_norank, Archaea_unclassified, and Nitrososphaera were the dominant genera in the four fertilization treatments (Fig. S3a,b). The composition of the AOA communities in the soils of the M and CK treatments was largely similar, and the predominant archaeal genera were Environmental_samples_norank (64.51% and 68.43%, respectively) and Archaea_unclassified (35.44% and 31.53%, respectively). The predominant archaea in the N1 and N2 treatment soils differed from those in the CK treatment soil. The abundance of Environmental_samples_norank in the N1 and N2 treatment soils was 77.11% and 84.29% lower, respectively, than that in the CK treatment, and the abundant archaeal genus Nitrososphaera was significant enrichment in N1 (23.39%) and N2 (46.60%) (Fig. 1a).
The predominant phyla in the AOB communities in the four treatments included the Proteobacteria, Bacteria_unclassified, and environmental_samples (Fig. S3c,d). The composition of the AOB communities in the M and CK treatment soils was largely similar, and the enrichment bacteria were Nitrosomonadales_unclassified (54.58% and 62.24%, respectively), and Bacteria_unclassified (7.25% and 7.20%, respectively). The predominant bacteria in the N1 and N2 treatment soils were significantly different from those in the CK treatment soils. The most abundant bacterial genus was Nitrosospira (N1: 77.57% and N2: 82.63%), followed by Bacteria_unclassified (N1: 15.90% and N2: 7.68%) (Fig. 1b). In the N1 and N2 treatment soils, the abundance of Nitrosospira was 1.20-fold higher, Bacteria_unclassified was 1.19-fold and 5.93% higher, respectively, compared to that in the CK treatment soils.
Correlations between soil properties and AOA and AOB community
Redundancy analysis (RDA) of the unweighted UniFrac distance between samples revealed a strong degree of clustering of established AOA communities in relation to the four treatments (Fig. 2). All samples were well separated along the PC1 axis for both AOA and AOB, which explained 82.87 and 90.44% of the total variation. Organic and inorganic fertilizer treatment were distinguished in two clusters: one cluster included the CK and M treatments, whereas the other cluster comprised N1 and N2 samples. Moreover, the N1 and N2 clusters resolved along the PC2 axis and explained 15.42% and 7.83% of the total variation for AOA and AOB.
RDA also showed that the variation in the AOA and AOB community structure could be explained by all seven of the soil properties. The AOA and AOB community of organic fertilizer were changed mainly by the soil pH. The inorganic fertilizer treatments changed by the different soil properties. The N2 treatment showed a positive correlation with the AP and TN content, and the N1 treatment showed a positive correlation with AN and AK. The mantel test showed (Fig. 3, Table S2) that the main contributor to the differences in AOA community structures was soil pH (r = 0.89, p < 0.05),, and the secondary contributors to the AOA community structure differences were the TN (r = 0.61, p < 0.05), NN (r = 0.40, p < 0.05) and AP (r = 0.40, p < 0.05) contents. The soil properties affected the AOB community structure according to the order (P < 0.05): pH (r = 0.87) > TN (r = 0.56) > AP (r = 0.33).
Pairwise comparisons of environmental factors are shown, with a color gradient denoting Spearman’s correlation coefficients. AOA and AOB community composition was related to each environmental factor by partial (geographic distance-corrected) Mantel tests. Edge width corresponds to the Mantel’s r statistic for the corresponding distance correlations, and edge color denotes the statistical significance.
Co-occurrence pattern of soil AOA and AOB
To evaluate the impact of the different fertilizer treatments on microbial associations, six networks at the OTU level were constructed for the AOA and AOB, respectively (Fig. 4). Overall, the co-occurrence of soil AOA communities was more complex than that of the AOB communities, and fertilizer application caused differences in the co-occurrence patterns of the AOA and AOB. The number of nodes for AOA in networks of the inorganic fertilizer group was higher by 15.09%, and the number of edges was higher by 25.98%, respectively, compared to those of the organic fertilizer group. The number of nodes for AOB in networks of the inorganic fertilizer group was higher by 5.41%, and the number of edges was lower by 11.56%. compared to those of the organic fertilizer group. The positive correlation coefficients also differed, and the inorganic fertilizer group had lower values than the organic fertilizer group for AOA but higher values than the organic fertilizer group for AOB. These results suggest that the amount of mutually beneficial cooperation in AOB communities increased under organic fertilizer group; however, there was an increased level of competition in AOA communities in organic fertilizer group.
Hub analysis further suggested that the AOA and AOB taxa differed greatly among the organic and inorganic fertilizer groups. We selected the top 10 microbial taxa for this study (Tables S3,S4). The diversity of both the hubs AOA and AOB increased in the inorganic fertilizer group than that in the organic fertilizer group. Most of the connected hub AOA and AOB in the organic fertilizer group were Thaumarchaeota and Nitrosospira. The connected hubs AOA and AOB in the inorganic fertilizer group included Thaumarchaeota, Nitrosospira, Nitrososphaera, and unranked Proteobacteria. The different key hub operational taxonomic units (OTUs) in both AOA and AOB taxa indicated that the inorganic and organic fertilizers changed the dependence of connected microbial species. The data on the network module class also supported these results, and the hub OTUs in the inorganic and organic fertilizer groups belonged to different module classes.
Microbial assembly process of AOA and AOB in soil
The obtained betaNTI value indicated that stochastic processes were crucial in shaping the AOA community assembly, and the relationships between the occurrence frequency of OTUs and their relative abundance variations were 69% and 80%. We acknowledge that the organic fertilizer negligibly increased the stochastic processes for the AOA community assembly and significantly increased the deterministic processes for the AOB community assembly compared to the inorganic fertilizers (Fig. 5). The NCM successfully estimated a large fraction of the relationship between the occurrence frequency of OTUs and their relative abundance variations (Fig. 6), with 89% and 79% of explained AOA community variance for the organic fertilizer group and inorganic fertilizer group, respectively. The Nm-value was higher for both AOA (Nm = 19,273) and AOB (Nm = 5731) with the inorganic fertilizer group than that of the organic fertilizer group (Nm = 17,844; Nm = 3288). These results indicate that inorganic fertilizer increased the species diffusion of AOA and AOB.
Neutral community model (NCM) of AOA and AOB community assembly. The predicted occurrence frequencies for inorganic fertilizer and organic fertilizer, and all representing soil AOA and AOB communities. The solid blue lines indicate the best fit to the NCM, and the dashed blue lines represent 95% confidence intervals around the model prediction. OTUs that occur more or less frequently than predicted by the NCM are shown in different colors. Nm indicates the metacommunity size times immigration, and R indicates the fit to the model.
Discussion
Changes in the abundance of the AOA and AOB
AOA are known to be more suitable than AOB in acidic soils, while AOB are more active in alkaline and neutral environments28. However, some studies have identified AOB as the dominant ammonia oxidizers in several acidic soils29, and play a major role in acidic soil nitrification30,31,32. Moreover, AOB can exhibit high nitrification rates under a soil pH of 333. From the correlation analysis, we established that soil pH was the main factor affecting the AOA and AOB amoA copy numbers, similar to the results of Li et al.34 and Singh et al.35. We speculate that the differences in the results are due to a variation in the relative abundances of AOA and AOB from fractional to several orders of magnitude depending on factors in addition to soil pH, including organic matter, nitrogen content and other environmental factor36. In our study, long-term fertilization with inorganic fertilizers reduced the growth of AOA but increased the growth of AOB in black soil compared to that with organic fertilizers. This results was similar to the obvious research, which indicated that showed that chemical fertilizers can decrease the diversity of the AOA community2,37. Organic fertilizer provides a variety of inorganic and organic nutrients, such as organic acids, for crops while fertilizing the soil, maintaining soil pH and improving the soil structure and ecosystem38,39. The VPA in our study showed that soil pH explained 78.09% and 60.58% and TN explained 60.58% and 53.09% of the observed changes in the AOA and AOB communities, respectively (Table 3). This suggests that the high soil N content and the relatively low pH of soils were the two main reasons leading to the differences in the AOA and AOB communities after inorganic fertilizer application in black soil. Increasing the soil pH and providing more complex nutrients (e.g., amino acid content) may be the reason for maintaining the soil AOA and AOB community after organic fertilizers application40.
A pot experiment showed that Nitrosospira (AOB) and Nitrososphaera (AOA) dominated absolutely in organic fertilizer treatments in which the soil pH was 7.8–8.413. In our study, Nitrosospira and Nitrososphaera were the dominant microorganisms in the soils of the inorganic fertilizer group, which had relatively low pH values (5.47–5.62), but not in the soils of the organic fertilizer group, which had higher pH values (6.48–6.59). This phenomenon is not only related to the physical and chemical properties of the soil but also the acid sensitivity, substrate utilization, and other characteristics of microbes. Previous research showed that Nitrosospira of AOB play a vital role in the nitrification process in acidic soil following both long-term inorganic and organic fertilization41. After isolation, some Nitrosospira spp. were possibly involved in the ammonium oxidation at low soil pH of 442. We speculate this as a reason for the abundance of Nitrosospira in the inorganic fertilizer treatment in our study. Advanced technology is required to isolate additional strains of AOB that cannot be cultivated in the experiment and confirm the above. In addition, Zhalnina et al.43 showed that the abundance of Nitrososphaera negatively correlates with the abundance of bradyrhizobia. The Nitrosomonadales, which are beneficial for plant growth and inhibition of pathogens causing root rot44,45, were abundant in the organic fertilizer in our study. Therefore, for the development of sustainable agriculture, we must thoroughly consider microbial changes and adjust the fertilization strategy, e.g., usage of organic fertilizers instead of chemical fertilizers, to avoid microbial disorders.
Changes in the co-occurrence pattern of the AOA and AOB
Co-occurrence patterns are helpful for evaluating changes in microbial community structure and provide insights into potential microbial interactions46. Banerjee et al.47 indicated that intensive agricultural production could decrease the network complexity of bacteria. The corresponding positive and negative correlation coefficients further showed that intraspecific competition for AOA and mutual benefit symbiosis for AOB were increased in the inorganic fertilizer treatments. Network analysis could define the niche space of microorganism communities for soil management48. Inorganic fertilizer increased the network complexity of AOA but decreased the network complexity of AOB that can be attributed to the high eco-physiological diversity and survival in a wider habitat of AOB13. Fortunately, we also found that chemical fertilizers can indeed significantly increase the niche width of AOB and negligibly increase the niche width of AOA compared with that by organic fertilizers (Fig. S4). Therefore, after inorganic fertilizer application, the AOB community occupied more ecological niches and consumed more resources, resulting in a decreasing connection. To increase the ability of competitiveness with AOB for more resources, the AOA community need more cooperation, resulting in the increasing connection after inorganic fertilizer. Moreover, the hub Nitrososphaera increased significantly in the network, and some important Proteobacteria were missing after inorganic fertilizer application. This shift might result the functional changes in the nitrogen cycle and microbial assembly process of the AOA and AOB.
Changes in microbial assembly process of the AOA and AOB
Compared to organic fertilizer, inorganic fertilizer affects the microbial assembly process of AOB more than AOA. Research has shown that when the pH decreases, ammonia (NH3) is converted into ammonium (NH4+), affecting the acquisition of AOB, thereby increasing the number and activity of AOB49. Rütting et al.50 indicated that the contribution to ammonia oxidation of AOB outcompeted AOA under higher ammonium supply. The results are in accordance with environmental data suggesting that AOA are mainly responsible for ammonia oxidation under more oligotrophic conditions, whereas AOB dominate under eutrophic conditions51. When the AOB community has sufficient resource to survive, the structure of the AOB community is not easily affected by soil physicochemical properties (e. g. NH4+); however, it is affected by the birth and death of microorganisms. Thus, the microbial assembly process of AOB changed from a deterministic process in organic fertilizer to a stochastic process in inorganic fertilizer. However, AOA possess much higher substrate affinities than the comammox or AOB counterparts52, resulting in the microbial assembly process of AOA being less susceptible to substrate changes from inorganic fertilizer application.
Assembly mechanism could be one of the key processes in shifting the microbial functions. When deterministic assembly dominates the assembly process, a higher diversity of the community would generally show better reactor performance, and when the stochasticity dominates the assembly process, the functional performance declines1. In our study, we found that inorganic fertilizer could highly increase aerobic ammonia oxidation, nitrification, and ureolysis process compared to that with organic fertilizer (Fig. S5). This might be the reason for organic fertilizer having more amoA genes of AOA but lower functional reads than inorganic fertilizer when they are under the stochastic assembly process.
Therefore, inorganic fertilizer changes the abundance of AOA and AOB and also affect the interaction and strengthens the nitrosation process of soil ammonia-oxidizing microorganism. Surely, additional nitrosation processes may lead to loss of nitrogen and contamination of groundwater caused by excessive application of inorganic fertilizers. There is still much work to be explored to develop sustainable agriculture by maintaining a dynamic balance between fertilizer application, crop yield, and ecological stability.
In conclusion, organic and inorganic fertilizer distinctly alter the abundance, co-occurrence pattern, and microbial assembly process of soil AOA and AOB differently. Inorganic fertilizer decreased the amoA copy numbers of AOA and increased the amoA copy numbers of AOB. At the same time, compared with organic fertilizer, inorganic fertilizer could imply an ecological imbalance among AOA and AOB communities in enriching Nitrososphaera and Nitrosospira and decreasing Nitrosomonadales. Changes in soil AOA and AOB community structure and amoA copy numbers were primarily due to shifts in the low pH and high NN and AP content by application of inorganic fertilizer. Moreover, inorganic fertilizer increased the complexity of the co-occurrence pattern of AOA but decreased the complexity pattern of AOB. Our results also suggest that the inorganic fertilizer has little effect on the microbial assembly process of AOA, whereas the application of inorganic fertilizer significantly increases the stochastic process of AOB. Overall, organic fertilizer has less disturbance for AOA and AOB communities by maintaining neutral soil pH and alleviate the accumulation of NO3−N content. This study provides critical data for understanding the influence of inorganic and organic fertilizers on the soil ammonia-oxidizing archaeal and bacterial ecosystem, which will provide robust evidence to facilitate reasonable farmland fertilization strategies.
Materials and methods
Experimental design
The trial site located in Harbin, Heilongjiang Province, China (N 45° 40′, E 126° 35′). The four fertilization treatments assessed were CK (no fertilizer); N1 (low inorganic fertilizer), N2 (high inorganic fertilizer), and M (organic fertilizer, horse manure). The inorganic fertilizers used were urea, calcium super phosphate, and ammonium hydrogen phosphate. The dose of horse manure as an organic fertilizer was approximately 18,600 kg manure/ha. Fertilizer application details are provided in Table S5. The dose of fertilizer in this study referred to local fertilization level in northeast china.
Black soil samples
We collected black soil samples from a 35-year positioning trial filed located in Harbin, Heilongjiang Province (N 45° 40′, E 126° 35′), which is one of the experimental stations belonging to China’s agricultural research system. During soybean harvest, we collected rhizosphere soil samples at 10 points by randomly extracting 20–25 bean individuals in each plot. Soil which loosely adhered to the roots were gently shaken off and the soil tightly adhered to roots were brushed with a brush, and mixing to obtain one composite soil sample. Five samples of each treatment and a total of 20 samples were collected, and all the soil samples were transported to the lab on ice. Some soil samples in each treatment were selected randomly and air-dried in the shade for analysis of physical and chemical properties. Some soil samples were randomly selected and stored at − 80 °C for analysis of microorganism.
Soil properties
The soil pH was measured using a pH meter, which the ratio of soil and water was 1:153. Kjeldahl method was used to detect the soil TN, and NO3−-N and NH4+-N content were measured by flow injection analysis54. Mo‐Sb colorimetric method was used to determine the soil AP level55. The soil AK content was evaluated by Flame photometry. Soil OM content was measured by the K2Cr2O7 capacitance method56.
Soybean yield
Dry weight of soybeans harvested from a 10 m2 area in the central part of each plot was determined to calculate the soybean yield57.
DNA extraction and qPCR
Total DNA of soil microorganism was extracted with a Power Soil DNA Isolation Kit (MOBIO Laboratories Inc., Carlsbad, CA, USA) and was stored at − 80 °C until further analysis. The AOB primers used were amoA-1F and amoA-2R58. The AOA primers used were Arch-amoA-F and Arch-amoA-R59. The ABI 7500 Real‐Time PCR Detection System was used to survey the number of amoA genes (Applied Biosystems, Waltham, MA, USA). The 20 μL PCR reaction system contained 10 μL UltraSYBR Mixture (CWBIO, China); 2 μL cDNA template of AOA or AOB; 0.2 μL forward and reverse primers (10 μM); and 7.6 μL ddH2O. The qPCR amplification cycle for AOA comprised the following: 94 °C 5 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 63 °C for 40 s, and extension at 72 °C for 40 s. The qPCR amplification cycle for AOB comprised the following: 94 °C 5 min, followed by 40 cycles of denaturation at 95 °C for 60 s, annealing at 53℃ for 60 s, and extension at 72 °C for 90 s.
Sequencing data processing
The primers used were the same as those used for qPCR. The total 25 μL PCR reaction system contained 40 ng of DNA template; 1 μL forward and reverse primers (10 μM); 0.25 μL Q5 high-fidelity DNA polymerase; 5.0 μL 5× High GC Buffer; 5.0 μL 5× Reaction Buffer; 0.5 μL dNTP (10 mM). The PCR amplification cycle comprised the following: 30 s at 98 °C, followed by 27 cycles of denaturation at 98 °C for 15 s, annealing at 58 °C/55 °C (AOA/AOB) for 30 s, and extension at 72 °C for 30 s/45 s (AOA/AOB), with a final elongation step for 7 min at 72 °C. The PCR products were used to purify and generate the amplicon libraries, following which the data were analyzed using QIIME pipeline version 1.8.0. A total of 255,338 and 322,885 sequences (more than 10,000 sequences per individual soil sample) for AOA and AOB, respectively, were obtained after quality trimming (Tables S3,S4). Operational taxonomic units (OTUs) were defined by clustering at 97% similarity. The α‐diversity of AOA and AOB was calculated using mothur60 and with the following four parameters as richness indices: Shannon and Simpson diversity, Chao 1 and the abundance‐based coverage estimator (ACE). The pyrosequencing data could be detected in the NCBI by SRA accession (PRJNA51207; SRX1034826).
Statistical analysis
Interactions between the AOA and AOB communities and environmental factors were analyzed using CANOCO 5.0 for redundancy analysis (RDA). The network analysis was conducted at the OTU level of AOA and AOB for all the total 20 sample, and visualized with Gephi (version 0.9.2). Totally, 158 archaeal and 89 bacterial OTUs were analyzed. We calculated all pair-wise spearman correlations between OTUs by SPSS 20. The Spearman’s correlations |r|> 0.8 and P value of less than 0.01 were retained61. Moreover, among all the samples, according the RDA analysis, the samples were divided into two group: CK and M; N1 and N2. Thus, the two group were selected for the sub-network analysis. Sub-networks were produced from the total network by preserving the presented nodes and edges62. The topological parameters of the network were also computed using Gephi (version 0.9.2). We adopted a neutral community model (NCM) to predict the relationship between OTU detection frequencies and their relative abundance across the wider metacommunity, which were performed using R (version 3.6.3) and the program presented by Chen et al.20. Moreover, the assembly processes of AOA and AOB communities were evaluated by calculating the nearest taxon index and beta nearest taxon index (betaNTI) using the “ses.mntd” function in a picante package63,64. The contribution was considered a stochastic process when |betaNTI|< 2, and the shifts in community composition were deterministic processes when |betaNTI|> 2. The retained resulting correlations were imported into the Gephi platform to obtain the topology property parameters for the network65. Functional prediction was analyzed through FAPROTAX (http://www.cloud.biomicroclass.com). Soil physical and chemical property data were analyzed using Microsoft Excel 2010 pro and SPSS version 20.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. This study complies with relevant institutional, national, and international guidelines and legislation. We declare that we have the appropriate permissions for collection of plant or seed specimens.
Data availability
The datasets used and analyzed during the current study available from the corresponding author on reasonable request.
References
Zhang, H. L. et al. Effect of straw and straw biochar on the community structure and diversity of ammonia-oxidizing bacteria and archaea in rice-wheat rotation ecosystems. Sci. Rep. 9(1), 9367 (2019).
Gao, D. W., Liu, F. Q., Li, L., Chen, C. H. & Liang, H. Diversity and community structure of ammonia oxidizers in a marsh wetland of the northeast China. Appl. Microbiol. Biotechnol. 102, 8561 (2018).
Leininger, S. et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 (2006).
Bai, Y. C. et al. Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8(5), 694 (2020).
Xie, W. Y. et al. Long-term effects of manure and chemical fertilizers on soil antibiotic resistome. Soil Biol. Biochem. 122, 111–119 (2018).
Thiele-Bruhn, S., Bloem, J., de Vries, F. T., Kalbitz, K. & Wagg, C. Linking soil biodiversity and agricultural soil management. Curr. Opin. Environ. Sustain. 4, 523–528 (2012).
Rousk, J. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351 (2010).
Strickland, M. S. & Rousk, J. Considering fungal: Bacterial dominance in soils—Methods, controls, and ecosystem implications. Soil Biol. Biochem. 42, 1385–1395 (2010).
Zhou, J. et al. Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil Biol. Biochem. 90, 42–51 (2015).
Zhou, J. et al. Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast China. Soil Biol. Biochem. 95, 135–143 (2016).
Yang, O. Y., Norton, J. M., Stark, J. M., Reeve, J. R. & Habteselassie, M. Y. Ammonia-oxidizing bacteria are more responsive than archaea to nitrogen source in an agricultural soil. Soil Biol. Biochem. 96, 4–15 (2016).
Jia, Z. J. et al. Evidence for niche differentiation of nitrifying communities in grassland soils after 44 years of different field fertilization scenarios. Pedosphere 30(1), 87–97 (2020).
Wessén, E., Nyberg, K., Jansson, J. K. & Hallin, S. Responses of bacterial and archaeal ammonia oxidizers to soil organic and fertilizer amendments under long-term management. Appl. Soil Ecol. 45(3), 193–200 (2010).
Shi, Y. L., Liu, X. R. & Zhang, Q. W. Effects of combined biochar and organic fertilizer on nitrous oxide fluxes and the related nitrifier and denitrifier communities in a saline-alkali soil. Sci. Total Environ. 686, 199–211 (2019).
Liang, M. X. et al. Soil fungal networks maintain local dominance of ectomycorrhizal trees. Nat. Commun. 11(1), 2636 (2020).
Barberán, A., Bates, S. T., Casamayor, E. O. & Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351 (2012).
Jones, C. M. & Hallin, S. Geospatial variation in co-occurrence networks of nitrifying microbial guilds. Mol. Ecol. 28(2), 293–306 (2019).
Tripathi, B. M. et al. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12(4), 1072–1083 (2018).
Zhou, J. et al. Consistent effects of nitrogen fertilization on soil bacterial communities in black soils for two crop seasons in China. Sci. Rep. 7, 3267 (2017).
Chen, W. D. et al. Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome 7(1), 1–16 (2019).
Hubbell, S. P. The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32) (Princeton University Press, 2011).
Silvertown, J. Plant coexistence and the niche. Trends Ecol. Evol. 19, 605–611 (2004).
Sengupta, A. et al. Spatial gradients in the characteristics of soil-carbon fractions are associated with abiotic features but not microbial communities. Biogeosciences 16, 3911–3928 (2019).
Zhao, P. et al. Deterministic processes dominate soil microbial community assembly in subalpine coniferous forests on the Loess Plateau. Peer J 7, e6746 (2019).
Feng, Y. et al. Two key features influencing community assembly processes at regional scale: Initial state and degree of change in environmental conditions. Mol. Ecol. 27, 5238–5251 (2018).
Shen, D., Qian, H., Liu, Y., Zhao, S. & Luo, X. Nitrifier community assembly and species co-existence in forest and meadow soils across four sites in a temperate to tropical region. Appl. Soil Ecol. 171, 104342 (2022).
Yang, X., You, L., Hu, H. & Chen, Y. Conversion of grassland to cropland altered soil nitrogen-related microbial communities at large scales. Sci. Total Environ. 816, 151645 (2022).
Zhang, L. M., Hu, H. W., Shen, J. P. & He, J. Z. Ammonia oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045 (2012).
Petersen, D. G. et al. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 14, 993–1008 (2012).
Lin, Y. et al. Ammonia-oxidizing bacteria play an important role in nitrification of acidic soils: A meta-analysis. Geoderma 404, 115395 (2021).
Lehtovirta-Morley, L. E., Stoecker, K., Vilcinskas, A., Prosser, J. I. & Nicol, G. W. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. PNAS 108(38), 15892–15897 (2011).
Hayatsu, M. et al. An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. ISME J. 11(5), 1130–1141 (2017).
Hayatsu, M. & Kosuge, N. Autotrophic nitrification in acid tea soils. Soil Sci. Plant Nutr. 39, 209–217 (1993).
Li, Y. Y., Chapman, S. J., Nicol, G. W. & Yao, H. Y. Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 116, 290–301 (2018).
Singh, H., Verma, A., Ansari, M. W. & Shukla, A. Physiological response of rice (Oryza sativa L.) genotypes to elevated nitrogen applied under field conditions. Plant Signal. Behav. 9, e29015 (2014).
Sun, R., Myrold, D. D., Wang, D. Z., Guo, X. S. & Chu, H. Y. AOA and AOB communities respond differently to changes of soil pH under long-term fertilization. Soil Ecol. Lett. 1, 126–135 (2019).
Shen, J. P., Zhang, L. M., Zhu, Y. G., Zhang, J. B. & He, J. Z. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ. Microbiol. 10, 1601–1611 (2008).
Zhou, C. et al. A new strategy for co-composting dairy manure with rice straw: Addition of different inocula at three stages of composting. Waste Manag. 40, 38–43 (2015).
Meng, F. Q., Qiao, Y. H., Wu, W. L., Smith, P. & Scott, S. Environmental impacts and production performances of organic agriculture in china: A monetary valuation. J. Environ. Manag. 188, 49–57 (2017).
Lin, Y. X. et al. Long-term manure application increases soil organic matter and aggregation, and alters microbial community structure and keystone taxa. Soil Biol. Biochem. 134, 187–196 (2019).
Lin, Y. et al. Nitrosospira cluster 8a plays a predominant role in the nitrification process of a subtropical Ultisol under long-term inorganic and organic fertilization. Appl. Environ. Microbiol. 84(18), e01031-e1118 (2018).
De Boer, W., Gunnewiek, P. K. & Laanbroek, H. J. Ammonium-oxidation at low pH by a chemolithotrophic bacterium belonging to the genus Nitrosospira. Soil Biol. Biochem. 27(2), 127–132 (1995).
Zhalnina, K. et al. Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front. Microbiol. 4, 104 (2013).
Wan, Y., Li, W., Wang, J. & Shi, X. Bacterial diversity and community in response to long-term nitrogen fertilization gradient in citrus orchard soils. Diversity 13(7), 282 (2021).
Hong, L. I. et al. Integrated analysis reveals an association between the rhizosphere microbiome and root rot of arecanut palm. Pedosphere 31(5), 725–735 (2021).
Qi, X. E. et al. Changes in alpine grassland type drive niche differentiation of nitrifying communities on the Qinghai Tibetan plateau. Eur. J. Soil Biol. 104, 103316 (2021).
Banerjee, S. et al. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736 (2019).
Freilich, M. A. et al. Species co-occurrence networks: Can they reveal trophic and non-trophic interactions in ecological communities? Ecology 99, 690–699 (2018).
Claros, J. et al. Effect of pH and HNO2 concentration on the activity of ammonia-oxidizing bacteria in a partial nitration reactor. Water Sci. Technol. 67(11), 2587–2594 (2013).
Rütting, T., Schleusner, P., Hink, L. & Prosser, J. I. The contribution of ammonia-oxidizing archaea and bacteria to gross nitrification under different substrate availability. Soil Biol. Biochem. 160, 108353 (2021).
French, E., Kozlowski, J. A. & Bollmann, A. Competition between ammonia-oxidizing archaea and bacteria from freshwater environments. Appl. Environ. Microbiol. 87(20), e01038–e01121 (2021).
Jung, M. Y. et al. Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 16(1), 272–283 (2022).
Li, X. Y. et al. Shifts of functional gene representation in wheat rhizosphere microbial communities under elevated ozone. ISME J. 7, 660–671 (2013).
Mulvaney, R. L. Nitrogen-inorganic forms. In Method of Soil Analysis. Part 3. Chemical Methods (ed. Sparks, D. L.) 1123 (American Society of Agronomy and Soil Science Society of America, 1996).
Olsen, S. R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate (United States Department of Agriculture, 1954).
Ding, J. L. et al. Effects of applying inorganic fertilizer and organic manure for 35 years on the structure and diversity of ammonia-oxidizing archaea communities in a Chinese Mollisols field. Microbiol. Open 9(1), e00942 (2020).
Ding, J. L. et al. Effect of 35 years inorganic fertilizer and manure amendment on structure of bacterial and archaeal communities in black soil of northeast China. Appl. Soil. Ecol. 105, 187–195 (2016).
McTavish, H., Fuchs, J. & Hooper, A. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175, 2436–2444 (1993).
Francis, C. A. et al. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. 102, 14683–14688 (2005).
Schloss, P. D., Gevers, D. & Westcott, S. L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6(12), e27310 (2011).
Fan, K. K. et al. Microbial resistance promotes plant production in a four-decade nutrient fertilization experiment. Soil Biol. Biochem. 141, 107679 (2020).
Assenov, Y. et al. Computing topological parameters of biological networks. Bioinformatics 24, 282–284 (2008).
Stegen, J. C., Lin, X. J., Konopka, A. E. & Fredrickson, J. K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6(9), 1653–1664 (2012).
Xun, W. B. et al. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 10(1), 1–10 (2019).
Delgado-Baquerizo, M. et al. Ecological drivers of soil microbial diversity and soil biological networks in the Southern Hemisphere. Ecology 99, 583–596 (2018).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFD1700200), the China Postdoctoral Science Foundation (2021M693450), the National Natural Science Foundation of China for Young Scholars (41807053), and the earmarked fund for Modern Agro-industry Technology Research System (CARS-04). We would like to thank Dr. Baoku Zhou and Prof. Baisuo Zhao of Chinese Academy of Agricultural Sciences for the help of fieldwork and data analysis.
Author information
Authors and Affiliations
Contributions
M.C.M., X.J. and J.L. designed the experiment. Y.B.Z. and M.C.M. analyzed all data and wrote the first draft of the manuscript. D.W.G. and M.Y. performed field experiment. F.M.C., L.L., J.Z., J.L.D. and J.L. edited the manuscript and checked the language. All authors contributed to manuscript revisions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, M., Zhao, Y., Jiang, X. et al. Fertilization altered co-occurrence patterns and microbial assembly process of ammonia-oxidizing microorganisms. Sci Rep 13, 8234 (2023). https://doi.org/10.1038/s41598-022-26293-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26293-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.