Abstract
The distribution of marine organisms is shaped by geographic distance and oceanographic features like currents. Among migratory species, individuals from multiple populations may share feeding habitats seasonally or across life stages. Here, we introduce a modification for many-to-many mixed stock models to include distance between breeding and foraging sites as an ecological covariate and evaluate how the composition of green turtle, Chelonia mydas, juvenile mixed stock aggregations changed in response to population growth over time. Our modified many-to-many model is more informative and generally tightens credible intervals over models that do not incorporate distance. Moreover, we identified a decrease in genetic diversity in a Florida nesting site and two juvenile aggregations. Mixed stock aggregations in central Florida have changed from multiple sources to fewer dominant source populations over the past ~ 20 years. We demonstrate that shifts in contributions from source populations to mixed stock aggregations are likely associated with nesting population growth. Furthermore, our results highlight the importance of long-term monitoring and the need for periodical reassessment of reproductive populations and juvenile aggregations. Understanding how mixed stock aggregations change over time and how different life stages are connected is fundamental for the development of successful conservation plans for imperiled species.
Similar content being viewed by others
Introduction
Dispersal shapes species' distributions and genetic structure; organisms dispersing into new areas may select suitable habitats based on factors such as availability1, temperature2, resources, and competition3,4. Among mobile organisms, some have defined home ranges and low dispersal, such as the maned sloth (Bradypus torquatus)5, while others, such as the saltwater crocodile (Crocodylus porosus), are distributed more broadly and travel hundreds of kilometers for food and reproduction6. Particularly among migratory organisms, individuals originating from multiple populations may share the same habitat and resources (e.g., fishes7, and whales8,9). These shared habitats can be occupied by individuals from different populations during a specific time of the year, as in gray whales (Eschrichtius robustus) that seasonally share foraging habitats in the Pacific Ocean8, or during multiple years as in juvenile sea turtle foraging aggregations10.
In organisms for which different populations share a habitat, mixed stock analysis (MSA) is a useful technique to understand genetic connectivity between source populations and mixed stock aggregations11. There are different approaches for mixed stock calculations, most commonly using maximum likelihood12,13 or Bayesian inference14,15,16. In addition, there are models designed to use haploid or single-parent inherited markers (e.g., mitochondrial DNA [mtDNA])14,17,18 and nuclear or diploid markers (e.g., microsatellites, allozymes, minisatellites, among others)12,13,15. Regardless of the approach, such methods usually compare genetic marker frequencies from mixed stock aggregations to known reproductive populations to estimate contributions to mixed stocks16. For mtDNA and similar markers, Bolker et al.14 introduced a model that considers the contributions from all available source populations to multiple mixed stocks simultaneously (e.g., many-to-many models). Many-to-many models allow robust and more realistic estimates of source populations than previous models that only accepted one mixed stock aggregation as a possible destination (e.g., many-to-one models14,16). Despite the important methodological advancements introduced by Bolker et al.14, the many-to-many model introduced in the R package ‘mixstock' was not designed to consider site-specific variables such as the distance between source and destination sites. Nishizawa et al.19 added a matrix of distances between source populations and mixed stocks as priors to the model as a means to account for distance in a many-to-many framework; however, it is unclear how their distance matrix is incorporated into the model.
Understanding how demographic parameters and dispersal patterns impact mixed stock composition is fundamental for implementing conservation plans for endangered and threatened species, especially if such variables change over time. Under scenarios where individuals are free and capable of moving in any direction, we would expect large source populations to contribute more individuals to mixed stocks than small populations. However, most organisms disperse between habitats according to biogeographical and resource constraints1,3,4. Also, temporal changes in source population sizes and stochastic events altering dispersal patterns can hamper our ability to characterize source populations for mixed stocks16. A mixed stock model implemented by Okuyama and Bolker18 weights model contribution estimates based on the size of each source population. Mixed stock models often require robust sampling from all sites under evaluation to obtain reliable estimates17, leading to combined datasets from previously published studies, regardless of the time period when samples were collected7,9,10,20,21,22,23. A concern with such an approach is that source populations are not necessarily constant over time24,25, just as haplotype frequencies in mixed stock aggregations might fluctuate26. Furthermore, the distance traveled by individuals from source populations to foraging aggregations is, in general, an important variable impacting dispersal27,28, and is often not considered in mixed stock assessments7,8,9,10,11,29 (but see19,20,21). Therefore, there is a need to evaluate the impact of temporal changes in reproductive population demographics on mixed stock aggregations and to develop models that can better account for the distance between breeding and mixed stock aggregations into many-to-many MSAs.
The green turtle (Chelonia mydas) is an ideal organism to evaluate how demographic variations and distance between source populations (rookeries) impact mixed stock aggregations. First, green turtle foraging aggregations are typically composed of individuals from multiple populations10,14,22,23,26,29. Second, in the past two decades, the number of nests in several green turtle rookeries in the Greater Caribbean and the western North Atlantic have increased at different rates24,25. Recently, van der Zee et al.22 suggested that changes in contributions observed in a juvenile mixed stock in Bonaire could be associated with a variation in the size of the source nesting populations. Third, green turtles leave nesting beaches as hatchlings and swim away from the coast to offshore habitats where they reside for a number of years30. Even though oceanic-stage green turtles are not complete passive drifters and may actively swim and orient31, there is substantial evidence suggesting marine turtle juvenile dispersal is also influenced by oceanographic currents, especially during the first few years of their life cycle32,33,34 (but see35). Therefore, we can approximate the distance traveled by individuals between rookeries and mixed stock areas by following the main marine currents connecting the different areas. Lastly, the east coast of central Florida, USA, hosts one of the largest nesting aggregations for green turtles in the western North Atlantic25 and several mixed stock aggregations36,37.
Here, we evaluate the impact of temporal changes in rookery sizes and in green turtle mixed stock aggregations in the Greater Caribbean and western North Atlantic while accounting for distance traveled between rookeries and mixed stock aggregations. To achieve these goals, we (i) modify the many-to-many mixed stock model to weight estimates based on the distance between rookeries and mixed stocks, and use the modified model to (ii) evaluate how variations in rookery sizes impacted MSA estimates over a two-decade period, and (iii) assess how MSA estimates change in response to variation in mixed stock haplotype frequencies and rookery size over the same period.
Methods
Study site and data collection
Adding to the available data on haplotype frequencies for rookeries and mixed stocks (Supplementary tables S1–S3), we collected data from nesting female green turtles in the Brevard County portion of the Archie Carr National Wildlife Refuge, in Melbourne Beach, Florida, USA (28.04° N, 80.55° W to 27.87° N, 80.45° W—hereafter referred to as “MB”)38. We sampled juveniles at two mixed stock foraging sites: the Indian River Lagoon about two kilometers south of the Sebastian Inlet (27.82° N, 80.43° W—“IRL”), and at Trident Basin at Port Canaveral (28.42° N, 80.59° W—“TRID”), both on the east coast of central Florida, USA36,37. All specimens used in this study were collected in accordance with animal care and use protocols approved by the Institutional Animal Care and Usage Committee at the University of Central Florida (IACUC 2020-04, 2020-18, 2020-138, and their predecessors). Skin and blood samples collection were conducted under permits MTP-231, NMFS 19508, and their predecessors.
We defined two sampling periods, “old” and “new”, within the rookery and mixed stock samples: for the rookery, samples collected before 2000 = MBold, and samples collected in 2016 to 2018 = MBnew. At the mixed stock sites, samples from 2003 to 2005 = IRLold and TRIDold, while samples from 2016 to 2018 = IRLnew and TRIDnew. Nesting female samples were assigned to a sampling period based on their first encounter, while juvenile mixed stock samples were assigned to a sampling period if any of the capture dates occurred during the years examined in this study. We recorded the standard straight carapace length (SCL) from the nuchal notch to the tip of the longest pygal scute when possible39. We extracted DNA from either skin or blood samples. Skin samples were collected using a 4-mm biopsy punch and stored in 95% ethanol at room temperature. Blood samples were collected from the dorsal cervical sinus into vials with sodium or lithium heparin, centrifuged to separate plasma, and red blood cells were frozen at −20 °C. For most of the blood samples collected from nesting females before the year 2000, a subset of the whole blood was also stored at room temperature in lysis buffer (100 mM Tris–HCl, 100 mM EDTA, 10 mM NaCl, 1% SDS, pH 8.0) using a 1:10 ratio of blood to buffer40.
Laboratory analyses
We extracted genomic DNA using either a Qiagen DNeasy blood and tissue kit following the manufacturer’s protocol or a Serapure Bead method with adaptations41,42. We used primers LCM1538243 and CM1643729 to amplify an 829 bp fragment of the mitochondrial control region (mtDNA). We used 20 µL polymerase chain reactions with final concentrations of 20 mM Tris HCl pH 8.4, 50 mM KCl, 0.25 mM of each dNTP, 1.5 mM MgCl2, 0.5 µM of each primer, 1 unit of Taq DNA polymerase, approximately 10 ng of genomic DNA, and water. We set up thermal cyclers to the following conditions: 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 57 °C for 30 s, 72 °C for 80 s; and a final extension at 72 °C for 10 min; holding at 10 °C. Samples with haplotype CM-A1.1 were screened at one additional locus using primers CM12751F and CM13064R following PCR and sequencing conditions described in Shamblin et al.23. We purified all PCR reactions using Exonuclease I (EN0581) and FastAP (EF0651) following the manufacturer’s protocol. Samples were sequenced in both directions through Sanger sequencing.
Data analyses
We edited, assembled, and aligned mtDNA sequences to reference haplotypes (829 bp) available from the Archie Carr Center for Sea Turtle Research database (https://accstr.ufl.edu/resources/mtdna-sequences/) using Geneious R844. We created a median-joining haplotype network using PopART v1.745, and calculated pairwise fixation indexes (FST), nucleotide (π) and haplotype (h) diversities using Arlequin v3.5.2.246. We used FST thresholds proposed by Wright47 to assess population differentiation. We compared genetic variation over time for each sampling site via analysis of molecular variance (AMOVA) with 10,000 permutations in Arlequin.
Mixed stock analysis
We modified the many-to-many mixed stock model, originally implemented in the R package ‘mixstock'14. The original model available in the ‘mixstock' package accepts a covariate to weight estimates obtained from haplotype frequency data by the relative size of each rookery14,18. Although rookery size is an important factor influencing contributions from rookeries to mixed stocks, the current model does not accept site-specific factors such as distance between rookery and mixed stock location, or main marine currents in between. To date, researchers need to input a matrix of values as priors into the many-to-many model in order to add the effect of distance on estimates19. Our assumption is that rookeries might have greater contributions to relatively closer mixed stocks than to distant ones. Similarly, dispersal from rookeries to some mixed stock aggregations can be facilitated by oceanographic conditions. Even though juveniles are capable of orienting and actively swimming in marine currents31, there is a greater chance for individuals to disperse to areas closer to where currents initially lead them than to other locations. Following this rationale, we modified the many-to-many model to weight the expected contributions by the scaled inverse distance (P) between each source population and mixed stock pair. The model we introduce here is:
where SourceContribution is the estimated contribution from each rookery based on haplotype frequencies, and SourceSize is the estimated size of each source population. The modified model differs from Bolker et al.14 only by the scaled inverse distance (P) matrix. The code and rationale for the base model with SourceContribution and SourceSize are described in Bolker et al.14 and Okuyama and Bolker18. See Supplementary Document S1 for details on our modifications. Here, we populated the matrix with values derived from the estimated inverse distances between each rookery and mixed stock by measuring the length of probable paths between sites using available marine currents as vectors for transport between rookeries and mixed stock aggregations (Fig. 1; Supplementary Tables S4 and S5). Scaled estimates of effective inverse distance (P) between each pair of mixed stock (i) and rookery (j) were calculated by
where D is the estimated distance from the rookery j to the mixed stock i. Given that a variety of factors may influence the direction and intensity of marine currents48, we considered two different scenarios (Scenarios 1 and 2) in which individuals may take different paths to move between sites (Fig. 1—our discussion focuses only on Scenario 1. See Supplementary Table S4 for distance scenario 2). Our goal was to introduce a tool to improve future mixed stock analysis, not necessarily to define dispersal patterns for green turtles.
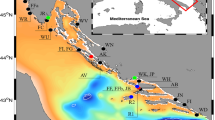
Probable routes used by juvenile green turtles to estimate the relative distances between rookeries and foraging areas; general current direction indicated by gray arrows. Blue triangles are rookeries, and green squares are mixed stock aggregation areas. Samples from MB are part of the CEFL rookery. Rookeries: AVES Aves Island, Venezuela, SURN Matapica and Galibi, Suriname, TORT Tortuguero, Costa Rica, MXQR Quintana Roo, Mexico, MXCA Campeche and Yucatán, Mexico, MXTV Tamaulipas and Veracruz, Mexico, SWCB Guanahacabibes Peninsula, Cuba, SOFL South Florida, United States, CEFL Central Florida, United States. Mixed stocks: BAR multiple areas, Barbados, BON Lac Bay, Bonaire, NIC Northeast Nicaragua, Nicaragua, BAH Southern Bahamas, The Bahamas, SWTX Southwest Texas, United States, NWFL Northwest Florida, United States, HISL Hutchinson Island, United States, RSBI Reef at Sebastian Inlet, United States, IRL Indian River Lagoon, United States, TRID Trident Basin, United States, CENC Central North Carolina, United States. Map generated in ArcMap 10.8.1 (https://www.esri.com), and arrows and labels were added in Adobe Illustrator 24.3 (https://www.adobe.com).
We used a short fragment of the mtDNA (491 bp) for our MSAs (Supplementary Tables S1–S3), which is contained within the longer (829 bp) fragment. We searched the literature for haplotype frequencies in other mixed stocks and rookeries within the western North Atlantic and Greater Caribbean (Fig. 1, Supplementary Tables S1–S3). We removed from our final dataset haplotypes found in mixed stock aggregations but not yet described in rookeries14, and considered only rookeries in the northwest Atlantic and the Greater Caribbean to reduce noise from unlikely contributors49. Though mixed stock data published by van der Zee et al.22 uses a timeframe slightly different than the one from IRL and TRID, we included their data in our dataset evaluating variations in haplotype frequencies to assess possible variations in other sites as well.
For rookery size we used a three-year average of the number of nests laid (Table 1), based on the best available data we had access to. We estimated rookery size for two time periods: historical (~ late 1990s) and recent (early 2010s). Finally, we included only rookeries in the western North Atlantic and Greater Caribbean for which data on the annual number of nests were available for both time periods considered (Table 1).
We ground-truthed our model to ensure that the modified model results were consistent with the original model in the ‘mixstock’ package when incorporating a matrix of ones (Supplementary Document S2). We also used a simulated dataset to compare the estimates from our modified model to the approach used by Nishizawa et al.19 and determine if results were similar. We compared estimates from the original many-to-many model (MSA1) to estimates from our modified model including a distance matrix (MSA2) to demonstrate how inclusion of a new covariate can impact MSA estimates. For the models described below (MSA3-MSA6) we populated the distance matrix with values from distance scenario 1 (Supplementary Table S4). To evaluate how changes in rookery sizes impacted MSA estimates, we combined all available data for each mixed stock into a single dataset (Supplementary Table S2) and created one model for each period: MSA3—“historical” source size, and MSA4—“recent” source size. Finally, to assess how contributions from rookeries changed over time based on mixed stocks haplotype frequencies and rookery sizes, we also built two models: MSA5—“old” sampling period and “historical” source size, and MSA6—“new” sampling period and “recent” source size. We considered samples from mixed stock BON22 to be from comparable timeframes (2006–07 and 2015–16) to IRL and TRID. Therefore, we added haplotype frequencies from BON to our "old" and "new" sampling periods in MSA5 and MSA6, and used the same haplotypic data from MSA3/MSA4 for all other mixed stocks (Supplementary Table S3). Even though the IRL, TRID, and BON are the only mixed stocks with data to answer our last goal, we included all other mixed stocks in models MSA5 and MSA6 to ensure estimates were more accurate. We used three chains for each model with a random starting point. We adjusted the number of iterations and burn-in period for models (Supplementary Table S6) to ensure chain convergence by checking the Gelman-Rubin shrink factor (< 1.08). To determine evidence of changes between models, we compared estimates by subtracting the posterior distributions for each estimated parameter between two models (e.g., MSA3 vs MSA4). The resulting distribution was used in the Test of Practical Equivalence implemented in the R package ‘bayestestR’56 against a null hypothesis (CI = 0.89, range = −0.05 to 0.05). In short, the Test of Practical Equivalence evaluates what proportion of the credible interval of the resulting distribution (i.e., 89% CI) that falls inside the range defined as the null (i.e., −0.05 to 0.05)56. We chose a credible interval of 89% based on small posterior distributions sample size (< 10,000)56,57. Finally, we used linear regressions to test if the mean estimates from models MSA3-MSA6 were correlated to the distance between rookeries and mixed stocks.
Results
Biometrics
We sampled a total of 200 turtles among the three locations and two time periods (Table 2). The mean SCL of first capture for nesting females in MBold was 100.9 cm (SD 5.1, range 93.4–114.1) and for MBnew was 97.8 cm (4.7, 90.5–108.6). For mixed stock samples, the mean size of first capture at IRLold was 46.8 cm (10.8, 29.5–68.6) and for IRLnew 48.1 cm (8.4, 32.4–66.7), while TRIDold was 29.5 cm (2.9, 23.4–39.2) and TRIDnew was 30.9 cm (4.3, 23.7–43).
Population structure
We identified four different haplotypes in MB (Table 2, Supplementary Fig. S1), including two samples with CM-A48.3 in MBold. This is the first time CM-A48.3 has been identified at a nesting site. The short-fragment version of this variant (CM-A48) had previously only been found in Cuba58. We identified haplotypes CM-A27.1 and CM-A28.1 for the first time in a juvenile foraging site on the east coast of Florida. Haplotypes CM-A3.1 and CM-A1.1 were the most frequent both in adult (41.7% and 41.7%) and in-water samples (45.8% and 25.0% in IRL, and 35% and 46.3% at TRID) for both “old” and “new” sampling periods (Table 2, Supplementary Fig. S1).
Results from AMOVAs to determine if genetic diversity changed over time per site indicate that most variation was observed within populations and not over time for all sites. However, we did find that among population variation was greater at the MB site, indicating greater change-over-time than found at the in-water sites (Supplementary Table S7). Haplotype (h) and nucleotide (π) diversities decreased for all sites over time. In MB, h decreased from 0.709 (SD = 0.047) before year 2000 to 0.567 (0.056) in 2016–2018, and π decreased from 2.252 × 10–3 (1.482 × 10–3) to 0.757 × 10–3 (0.693 × 10–3). For mixed stock aggregations, in the IRL h varied from 0.8 (0.049) in 2003–2005 to 0.65 (0.063) in 2016–2018 and π from 3.526 × 10–3 (2.112 × 10–3) to 2.899 × 10–3 (1.794 × 10–3), while in TRID h went from 0.734 (0.05) to 0.553 (0.068) and π from 2.451 × 10–3 (1.566 × 10–3) to 1.13 × 10–3 (0.885 × 10–3). The TRID mixed stock saw the highest reduction in haplotype diversity (from 0.732 to 0.553). Despite variation in haplotype and nucleotide diversities, pairwise FST comparisons did not indicate significant variation within sites over time (Table 3). The only differences in FST were between IRLold and both MB sampling periods, between IRLnew and MBold, and between TRIDnew and both IRL sampling periods. We found no evidence of structuring between sampling timeframes for each site. For rookery data, we also grouped samples with previous studies for the models evaluating changes in haplotype frequencies (MB is part of the CEFL rookery—Supplementary Table S1).
Distance matrix
Comparing the estimates obtained using our modified model and the approach used by Nishizawa et al.19 we found that our modified model consistently provided estimates with narrower credible intervals than adding distances as priors (Supplementary Fig. S2). Regarding the inclusion of a distance matrix to a many-to-many approach, estimates from rookery SWCB to the TRID mixed stock and from MXQR, SURN, and AVES to both mixed stocks remained essentially the same between MSA1 and MSA2 (Fig. 2). However, adding the distance matrix in MSA2 made the credible intervals wider from CEFL, MXCA, MXTV, TORT, and SWCB to the IRL mixed stock, and from MXCA and TORT to the TRID estimates. In contrast, credible intervals were narrower from SOFL to the IRL mixed stock, and from CEFL, SOFL, and MXTV to the IRL. We found a weak relationship between values populated into the distance matrix and rookeries contribution estimates for models MSA4-MSA6 (Supplementary Fig. S3).
Impact of incorporating a distance matrix into many-to-many mixed stock models. Solid points represent mean estimates and vertical bars 95% credible intervals. No Distance Matrix (MSA1) = standard many-to-many model from package ‘mixstock’; Distance Matrix (MSA2) = modified many-to-many model with a distance matrix. See Fig. 1 for site abbreviations.
Effect of rookery size on mixed stocks
All rookeries showed an increase in the average number of nests per season from historical to recent time periods (Table 1). However, given the different rates of increase, the relative contribution from each rookery (the number of nests divided by the total number of nests in all rookeries for each time period) changed over time. For AVES and SWCB, their relative proportion remained virtually unchanged (~ 1.5% and ~ 0.14% respectively). On the other hand, TORT had the largest increase in absolute numbers (over 42,000 annually), but its proportion decreased from 87.30 to 63.72%. For CEFL, SOFL, MWQR, MXCA, MXTV, and SURN there was an increase in their relative proportion at similar rates (3.09–4.93%).
We found little evidence of changes in contributions to mixed stocks as a response to changes in rookery sizes alone (MSA3 vs MSA4—Fig. 3). Some mixed stocks analyzed have a single main contributing source population: MXTV is the main contributor to TRID and SWTX, SURN is the main contributor to BAR, and TORT appears as the main contributor to BON, HISL, BAH, and NIC. Contributions to IRL, CENC, RSBI, and NWFL appear to come from multiple sources without a clear single origin. Considering the rookery-centric estimates (Supplementary Fig. S4), individuals from most rookeries disperse similarly among the mixed stocks analyzed (overall mean estimate 8.33%, SD 8.25%). Individuals from TORT disperse mainly to NIC, followed by other mixed stock(s) not present in this analysis (UNK). Main destinations for individuals originating from the SURN rookery were BAR, NIC, and UNK. For both TORT and SURN, there is great uncertainty regarding the main mixed stock destinations. Complete results for models using distance scenario 2 are available in Supplementary Tables S8 and S9.
Mixed stock-centric estimates comparing different source sizes. Filled dots represent the mean estimate, and vertical bars 95% credible intervals. MSA3 “historical” source size. MSA4 “recent” source size. Asterisk: rookeries with evidence of a difference in contribution in response to source size variation. See Fig. 1 for site abbreviations.
Haplotype and source size variation
For this objective, we present only the results for mixed stocks with data available for the two sampling periods and the corresponding rookery sizes: IRL, TRID, and BON (MSA5 and MSA6, Fig. 4—Supplementary Tables S3 and S10 for complete results). The impact of changes in source size on the broader mixed stock estimates was established in the previous section (models MSA3 and MSA4). For TRID, we found evidence of an increase in the proportion of individuals from MXTV, and for BON there was a decrease in contributions from SURN. Also, for the IRL mixed stock, recent years have narrower credible intervals and lower mean estimates for rookeries CEFL, SOFL, MXQR, SWCB, SURN, and AVES, while wider credible intervals were observed for MXCA and TORT. For TRID, we observed the same pattern of narrower credible intervals for all rookeries except for MXTV. Finally, for BON, recent years appear with tighter credible intervals for SURN and AVES, while for CEFL, SOFL, MXQR, MXCA, MXTV, and TORT we see wider credible intervals (Fig. 4). Source-centric estimates for MSA5 and MSA6 indicate no changes in destination of individuals from all rookeries over time (Supplementary Fig. S5).
Mixed stock-centric estimates with different sampling events and varying source sizes. Circles represent the mean estimate, and vertical bars 95% credible intervals. MSA5 “old” mixed stock sampling period and “historical” source size. MSA6 “new” mixed stock sampling period and “recent” source size. Asterisk: rookeries with evidence of a difference in contribution in response to source size variation. See Fig. 1 for site abbreviations.
Discussion
Our study introduces an important advancement for mixed stock analysis: a more informative and ecologically meaningful model incorporating a matrix of site to site-specific weighted inverse distances. We demonstrate how demographic variations in source populations and temporal changes in haplotype frequencies in mixed stocks aggregations can impact MSA estimates. Also, we show how understanding dispersal patterns and connectivity between sites is crucial for management of migratory organisms. Our analyses indicate how the stock composition of juvenile aggregations of green turtles in east central Florida have changed over a 13-year period, simultaneous to a population growth on several source nesting populations. Furthermore, we clearly demonstrate the importance of long-term monitoring and periodic reassessment of both breeding and juvenile aggregations.
For the juvenile IRL and TRID mixed stocks, the mean SCL from individuals sampled in our study was comparable to the mean sizes previously reported for both sites37,59. For the rookery MB, the mean size and observed reduction in mean SCL among nesting females are consistent with a trend recently described for this populations60. The pairwise comparison between sites (Table 3) corroborates our decision to treat IRL and TRID as separate mixed stocks. Also, FST indicates a greater genetic differentiation between MB and the IRL mixed stock, suggesting it is not mostly composed of individuals from MB despite geographical proximity, supporting our assumption that the distance between MB and IRL is much greater than what a straight line between these sites suggests (Supplementary Tables S4-S5). Several green sea turtle nesting sites in the western North Atlantic and Greater Caribbean have increased in both estimated abundance and number of nests laid24, including the MB nesting aggregation25. Female sea turtles are known for reproductive natal philopatric behavior10. Given that the genetic marker we used (mtDNA) is both haplotypic and maternally inherited, a reduction in haplotype diversity in reproductive populations would be expected given the reduced effective size associated with mtDNA, especially for historically bottlenecked populations. Regarding in-water aggregations, the observed reductions in h and π could be a consequence of changes in the main contributors to each mixed stock (Fig. 4), with a general homogenization of the genetic pool. Similarly, a recent study on a mixed stock in Lac Bay, Bonaire, found a reduction in nucleotide diversity but no clear change in haplotype diversity over 9 years22. However, van der Zee et al.22 amplified only the short mtDNA fragment, which could reduce their ability to detect variations. Regardless, results from our analysis indicate a predominance of a single contributor in BON in recent years instead of two from the "old" sampling period (Fig. 4), supporting the van der Zee et al.22 hypothesis of changes in contributors over time. Even though it is unlikely to be observed on all sites simultaneously, after splitting our dataset for IRL and TRID into two sampling periods, we cannot discard the possibility that these reductions are due to small sample sizes. The reduction in sample size for IRL, TRID, and BON mixed stocks in MSA5/MSA6 compared to MSA3/MSA4 could help explain the increased uncertainty around the estimates (Fig. 4). Additionally, we acknowledge that our results are a snapshot in time and encompass less than one generation-time for this species; undetected complex ecological processes might be underway61. Future studies with a larger sample size from a single mixed stock and time period could try to address this concern using a resampling approach (e.g., jackknife-based method) to identify how sampling might affect MSA estimates.
We identified variations on the width of credible intervals between our modified model and the original model in the ‘mixstock’ package (MSA1 vs MSA2). Even though we did not specifically test possible causes for variation in credible intervals after the inclusion of the distance matrix, we suspect it could be related to values in the distance matrix that do not match estimates from haplotype frequencies (e.g., haplotype frequencies indicate small contribution from one rookery while the value in the distance matrix suggest higher contribution from the same source). Mixed stocks and/or rookeries with small sample sizes could be more impacted by such variations.
We found no clear evidence of changes in contributors to mixed stocks when considering variation in rookery size alone (MSA3 and MSA4; Fig. 3). Previous studies report little or no difference in estimates when comparing models with rookery size versus models with an uninformative covariate (i.e., equal value to all rookeries) while using a many-to-one framework21,23. This is not an unexpected result as MSA estimates are mostly derived from genetic markers16, and the weight provided by covariates might not be enough to change estimates. However, we found evidence of variations in contributions when the haplotypic variation was considered along with rookery size variation (MSA5 and MSA6; Fig. 4). Though, we did not test a model with varying haplotype frequencies and constant rookery size, as rookery sizes did change over time this would be an unrealistic scenario and we could not tease these changes apart. Therefore, we cannot determine if the observed fluctuation in haplotype frequencies (and rookery contribution estimates) was caused by changes in the influx of individuals from source populations to mixed stocks or by variation in source population sizes because both possibilities are intrinsically dependent on one another.
The main contributors to mixed stocks from models MSA3 and MSA4 were partially different from previous analyses in the Atlantic Ocean and Greater Caribbean14,22,62,63,64,65,66,67,68,69, which could be explained by substantial differences between our dataset and the different datasets and models used by previous studies. However, results from MSA5 and MSA6 corroborate findings from studies that identified fluctuations in contributions over time in response to changes in haplotype frequencies in mixed stocks22,26. An assumption of mixed stock models is that all source populations are represented and adequately sampled16—an assumption that will rarely be met. Engstrom et al.49 suggest not including unlikely contributors to mixed stock models to reduce noise, a decision we also made. However, researchers may have different thresholds to define an unlikely contributor, therefore, this decision becomes arbitrary. Comparing estimates among studies is difficult as new areas are added and more samples are sequenced. Furthermore, our modified model uses effective distance to weight estimates; this adds an extra layer of differentiation among studies, making direct study comparisons even harder. Regardless of agreement (or lack of agreement) between our results and previous studies, we believe that future assessments can improve biological meaning if mixed stocks and rookeries are periodically reassessed for haplotype frequencies.
An increase in juvenile abundance following reproductive population growth and increased number of nests laid is a reasonable expectation. This expectation depends on the fitness of reproductive individuals, the hatching success of the nests laid, and survival and recruitment rates for juveniles. However, Bjorndal et al.61 found no correlation between increased number of nests at Tortuguero, Costa Rica, the putative main stock of origin for the mixed stock, and variations in the abundance of green turtles at Union Creek, The Bahamas. One hypothesis was that Union Creek was near carrying capacity for green turtles, and abundance would remain stable over time61. Our models corroborate their findings, showing TORT as the main contributor (Fig. 3) despite the reduction in TORT’s size in relationship to the other source populations in the region (Table 1). The stability of contributions to BAH could be an indication that the carrying capacity hypothesis is still a valid option for Union Creek. Similarly, Long et al.59 attributed a decrease in green turtle abundance in the IRL mixed stock between 2001 and 2018 to a general decrease in habitat quality, despite the increased abundance in rookeries. It is possible that juvenile abundance increased in other mixed stock aggregations and that the observations in the IRL and BAH mixed stocks59,61 are isolated cases. However, a study with green turtles from MB identified a decrease in nesting females’ mean size and size at maturity over the past decades, especially after the late 1990s60. One of the explanations for a decrease in nesting female body size is reduced juvenile mass growth rate70, which, ultimately, could lead to overall reduced reproductive fitness in rookeries. At least for leatherback sea turtles (Dermochelys coriacea), reproductive fitness can be impacted by maternal health parameters71. Interestingly, these data are supported by our genetic analyses that found little change between in-water sites over time, but greater change among time periods for the nesting beach site.
Mixed stock analysis using either the number of nests or the number of nesting females as a proxy for source size should correct estimates by emergence success (total number of hatchlings emerged divided by the total number of hatched eggs in a clutch). Emergence success can vary among seasons, rookeries, species, and can be affected by maternal health and environmental factors71,72. To ensure future mixed stock analyses benefit from more informative rookery sizes, we urge researchers to report the number of nests, hatching success, and emerging success, as well as other basic reproductive parameters from nesting populations. We second the call by Shamblin et al.73 for broad use of longer fragments of mtDNA in reassessments of rookeries that have been only evaluated using the short fragment, and especially, that new studies refrain from sequencing the short fragments only. The development of new diagnostic markers using whole mitogenomic sequences29, or a combination of mtDNA with other markers (e.g., nuclear microsattelites), to increase discrimination between rookeries is essential for our understanding of sea turtle evolution and dispersal patterns. Future population and species assessments will benefit from better and more refined genetic information.
Understanding dispersal and connectivity among habitats and across life stages is fundamental for species’ conservation. The main feature introduced by our modified model is the capacity for researchers to more easily consider variables that are specific to each pair of source populations and mixed stocks in a many-to-many framework. Prior to our modified model, studies incorporating particle dispersal probabilities or distance between sites often weighted MSA estimates using a many-to-one model framework because the probabilities from a source will differ to each mixed stock, and estimates from multiple mixed stock models need to be combined for a regional overview20,21,23,33. Many-to-many models provide estimates with narrower credible intervals than many-to-one models when analyzing the same dataset14,63,74. Our modified model usually provided narrower credible intervals than the approach introduced by Nishizawa et al.19 on a many-to-many framework. Site-specific probability matrices that incorporate complex variables such as particle dispersal model estimates will enable researchers to consider multiple cohorts, variation within and among seasons, and multiple variables that can impact oceanographic currents34.
Our modified many-to-many mixed stock model can incorporate new variables to make models more informative. More importantly, by incorporating distance between rookeries and mixed stocks, or particle dispersal probabilities, models we can better account for unlikely source populations, allowing more realistic estimates of rookery contributions to mixed stocks for robust ocean basin analyses. The short-fragment mtDNA markers used for MSA lack the resolution needed to differentiate between several rookeries29. As mixed stock model estimates are mainly derived from haplotype frequencies14,16, under scenarios where the genetic marker used is unable to differentiate populations, covariates can help improve model accuracy. The source code and example script for incorporating the site-specific matrix is available in Supplementary Document S1, and we encourage others to use this approach to incorporate distances, transport probabilities, or any other metric that scales the contributions from each rookery to each mixed stock. Contribution estimates from such models will be more ecologically meaningful and more accurate. Further, we highlight the importance of long-term monitoring and periodic reassessment of mixed stock aggregations regarding stocks of origin, abundance, health status, and other population parameters. We also emphasize the importance of periodical reassessment of haplotype frequencies at rookeries, as well as basic demographic and reproductive parameters. For migratory endangered species such as sea turtles, broad analyses considering multiple rookeries within or among ocean basins with more informative estimates are critical for understanding dispersal, connectivity, and evolution. Understanding how the composition of mixed stock aggregations shift over time is fundamental for the development of successful conservation plans for endangered and threatened species.
Data availability
Data used in mixed stock analyses and code for the modified model with detailed instructions are available as supplementary data.
References
MacPherson, E. Ontogenetic shifts in habitat use and aggregation in juvenile sparid fishes. J. Exp. Mar. Bio. Ecol. 220, 127–150 (1998).
Freitas, C., Olsen, E. M., Knutsen, H., Albretsen, J. & Moland, E. Temperature-associated habitat selection in a cold-water marine fish. J. Anim. Ecol. 85, 628–637 (2016).
Michelot, C. et al. Seasonal variation in coastal marine habitat use by the European shag: Insights from fine scale habitat selection modeling and diet. Deep. Res. Part II Top. Stud. Oceanogr. 141, 224–236 (2017).
Davoren, G. K., Montevecchi, W. A. & Anderson, J. T. Distributional patterns of a marine bird and its prey: Habitat selection based on prey and conspecific behaviour. Mar. Ecol. Prog. Ser. 256, 229–242 (2003).
Chiarello, A. G. et al. A translocation experiment for the conservation of maned sloths, Bradypus torquatus (Xenarthra, Bradypodidae). Biol. Conserv. 118, 421–430 (2004).
Fukuda, Y. et al. Environmental resistance and habitat quality influence dispersal of the saltwater crocodile. Mol. Ecol. 31, 1076–1092 (2022).
O’Leary, S. J., Dunton, K. J., King, T. L., Frisk, M. G. & Chapman, D. D. Genetic diversity and effective size of Atlantic sturgeon, Acipenser oxyrhinchus oxyrhinchus river spawning populations estimated from the microsatellite genotypes of marine-captured juveniles. Conserv. Genet. 15, 1173–1181 (2014).
Brüniche-Olsen, A. et al. Genetic data reveal mixed-stock aggregations of gray whales in the North Pacific Ocean. Biol. Lett. 14, 1–4 (2018).
Carroll, E. L. et al. Genetic diversity and connectivity of southern right whales (Eubalaena australis) found in the Brazil and Chile-Peru wintering grounds and the South Georgia (Islas Georgias Del Sur) feeding ground. J. Hered. 111, 263–276 (2020).
Bowen, A. B. W. et al. Origin of hawksbill turtles in a Caribbean feeding area as indicated by genetic markers. Ecol. Appl. 6, 566–572 (1996).
Paxton, K. L., Yau, M., Moore, F. R. & Irwin, D. E. Differential migratory timing of western populations of Wilson’s Warbler (Cardellina pusilla) revealed by mitochondrial DNA and stable isotopes. Auk 130, 689–698 (2013).
Anderson, E. C., Waples, R. S. & Kalinowski, S. T. An improved method for predicting the accuracy of genetic stock identification. Can. J. Fish. Aquat. Sci. 65, 1475–1486 (2008).
Debevec, E. M. SPAM (version 3.2): Statistics program for analyzing mixtures. J. Hered. 91, 509–511 (2000).
Bolker, B. M., Okuyama, T., Bjorndal, K. A. & Bolten, A. B. Incorporating multiple mixed stocks in mixed stock analysis: ‘Many-to-many’ analyses. Mol. Ecol. 16, 685–695 (2007).
Neaves, P. I., Wallace, C. G., Candy, J. R. & Beacham, T. D. CBayes: Computer Program for Mixed Stock Analysis of Allelic Data. Free Program Distributed by the Authors Over the Internet. at (2005).
Pella, J. & Masuda, M. Bayesian methods for analysis of stock mixtures from genetic characters. Fish. Bull. 99, 151–167 (2001).
Bolker, B., Okuyama, T., Bjorndal, K. A. & Bolten, A. B. Sea turtle stock estimation using genetic markers: Accounting for sampling error of rare genotypes. Ecol. Appl. 13, 763–775 (2003).
Okuyama, T. & Bolker, B. M. Combining genetic and ecological data to estimate sea turtle origins. Ecol. Appl. 15, 315–325 (2005).
Nishizawa, H. et al. Composition of green turtle feeding aggregations along the Japanese archipelago: Implications for changes in composition with current flow. Mar. Biol. 160, 2671–2685 (2013).
Naro-Maciel, E. et al. Predicting connectivity of green turtles at Palmyra Atoll, central Pacific: A focus on mtDNA and dispersal modelling. J. R. Soc. Interface 11, 20130888 (2014).
Proietti, M. C. et al. Green turtle Chelonia mydas mixed stocks in the western South Atlantic, as revealed by mtDNA haplotypes and drifter trajectories. Mar. Ecol. Prog. Ser. 447, 195–209 (2012).
van der Zee, J. P. et al. Population recovery changes population composition at a major southern Caribbean juvenile developmental habitat for the green turtle, Chelonia mydas. Sci. Rep. 9, 1–11 (2019).
Shamblin, B. M. et al. Mexican origins for the Texas green turtle foraging aggregation: A cautionary tale of incomplete baselines and poor marker resolution. J. Exp. Mar. Bio. Ecol. 488, 111–120 (2017).
Seminoff, J. A. et al. Status Review of the Green Turtle (Chelonia mydas) Under the Endangered Species Act. (NOAA Technical Memorandum, NOAA-NMFS-SWFSC, 2015).
Chaloupka, M. et al. Encouraging outlook for recovery of a once severely exploited marine megaherbivore. Glob. Ecol. Biogeogr. 17, 297–304 (2008).
Bjorndal, K. A. & Bolten, A. B. Annual variation in source contributions to a mixed stock: Implications for quantifying connectivity. Mol. Ecol. 17, 2185–2193 (2008).
Roland, J., Keyghobadi, N. & Fownes, S. Alpine Parnassius butterfly dispersal: Effects of landscape and population size. Ecology 81, 1642–1653 (2000).
Vanschoenwinkel, B., De Vries, C., Seaman, M. & Brendonck, L. The role of metacommunity processes in shaping invertebrate rock pool communities along a dispersal gradient. Oikos 116, 1255–1266 (2007).
Shamblin, B. M. et al. Mitogenomic sequences better resolve stock structure of southern Greater Caribbean green turtle rookeries. Mol. Ecol. 21, 2330–2340 (2012).
Witherington, B., Hirama, S. & Hardy, R. Young sea turtles of the pelagic Sargassum-dominated drift community: Habitat use, population density, and threats. Mar. Ecol. Prog. Ser. 463, 1–22 (2012).
Putman, N. F. & Mansfield, K. L. Direct evidence of swimming demonstrates active dispersal in the sea turtle ‘lost years’. Curr. Biol. 25, 1221–1227 (2015).
Mansfield, K. L., Wyneken, J. & Luo, J. First Atlantic satellite tracks of ‘lost years’ green turtles support the importance of the Sargasso Sea as a sea turtle nursery. Proc. R. Soc. B Biol. Sci. 288, 20210057 (2021).
Putman, N. F. et al. Predicted distributions and abundances of the sea turtle ‘lost years’ in the western North Atlantic Ocean. Ecography (Cop.) 43, 506–517 (2020).
Putman, N. F. & Naro-Maciel, E. Finding the ‘lost years’ in green turtles: Insights from ocean circulation models and genetic analysis. Proc. R. Soc. B Biol. Sci. 280, 20131468 (2013).
Naro-Maciel, E., Hart, K. M., Cruciata, R. & Putman, N. F. DNA and dispersal models highlight constrained connectivity in a migratory marine megavertebrate. Ecography (Cop.) 40, 586–597 (2017).
Ehrhart, L. M., Redfoot, W. E. & Bagley, D. A. Marine turtles of the central region of the Indian River Lagoon system, Florida. Florida Sci. 70, 415–434 (2007).
Redfoot, W. & Ehrhart, L. Trends in size class distribution, recaptures, and abundance of juvenile green turtles (Chelonia mydas) utilizing a rock riprap lined embayment at Port Canaveral, Florida, USA, as developmental habitat. Chelonian Conserv. Biol. 12, 252–261 (2013).
Ehrhart, L., Redfoot, W., Bagley, D. & Mansfield, K. Long-term trends in loggerhead (Caretta caretta) nesting and reproductive success at an important western Atlantic rookery. Chelonian Conserv. Biol. 13, 173–181 (2014).
Bolten, A. B. Techniques for measuring sea turtles. in Research and Management Techniques for the Conservation of Sea Turtles. (eds. Eckert, K. L., Bjorndal, K. A., Abreu-Grobois, F. A. & Donnelly, M.). 1–5 (1999).
Bagley, D. A. Characterizing Juvenile Green Turtles, (Chelonia mydas), from Three East Central Florida Developmental Habitats. (University of Central Florida, 2003).
Rohland, N. & Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012).
Faircloth, B. & Glenn, T. Preparation of an AMPure XP Substitute. AKA Serapure https://doi.org/10.6079/J9MW2F26 (2016).
Abreu-Grobois, F. A. et al. New mtDNA Dloop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analyses. in Proceedings of the 26th Annual Symposium on Sea Turtle Biology. 179 (International Sea Turtle Society, 2006).
Kearse, M. et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Leigh, J. W. & Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Wright, S. Evolution and the Genetics of Populations. Vol. 4. Variability Within and Among Natural Populations. (University of Chicago Press, 1978).
Hays, G. C. Ocean currents and marine life. Curr. Biol. 27, R470–R473 (2017).
Engstrom, T. N., Meylan, P. A. & Meylan, A. B. Origin of juvenile loggerhead turtles (Caretta caretta) in a tropical developmental habitat in Caribbean Panamá. Anim. Conserv. 5, 125–133 (2002).
Florida Fish and Wildlife Conservation Commission-Fish and Wildlife Research Institute, F. W. C. F. W. R. I. Index Nesting Beach Survey (INBS). (2021).
Cuevas Flores, E. A., Guzmán Hernández, V., Guerra Santos, J. J. & Rivas Hernández, G. A. El uso del Conocimiento de las Tortugas Marinas Como Herramienta para la Restauración de sus Poblaciones y Hábitats Asociados. (Universidad Autónoma del Carmen, 2019).
Pineda, O. G. & Rocha, A. R. B. Las Tortugas Marinas en México: Logros y Perspectivas para su Conservación. (CONANP, 2016).
Varela, R. G., Quílez, G. Z. & Harrison, E. Report on the 2014 Green Turtle Program at Tortuguero, Costa Rica. (2015).
Azanza Ricardo, J. et al. Nesting ecology of Chelonia mydas (Testudines: Cheloniidae) on the Guanahacabibes Peninsula. Cuba. Rev. Biol. Trop. 61, 1935–1945 (2013).
Nalovic, M. A. et al. Sea Turtles in the North Atlantic & Wider Caribbean Region. (2020).
Makowski, D., Ben-Shachar, M. & Lüdecke, D. bayestestR: Describing effects and their uncertainty, existence and significance within the Bayesian framework. J. Open Source Softw. 4, 1541 (2019).
Kruschke, J. K. Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan. https://doi.org/10.1016/B978-0-12-405888-0.09999-2 (Academic Press, 2015).
Ruiz-Urquiola, A. et al. Population genetic structure of greater Caribbean green turtles (Chelonia mydas) based on mitochondrial DNA sequences, with an emphasis on rookeries from southwestern Cuba. Rev. Investig. Mar. 31, 33–52 (2010).
Long, C. A. et al. Incongruent long-term trends of a marine consumer and primary producers in a habitat affected by nutrient pollution. Ecosphere 12, e03553 (2021).
Phillips, K. F., Stahelin, G. D., Chabot, R. M. & Mansfield, K. L. Long-term trends in marine turtle size at maturity at an important Atlantic rookery. Ecosphere 12, 7 (2021).
Bjorndal, K. A., Bolten, A. B. & Chaloupka, M. Y. Evaluating trends in abundance of immature green turtles, Chelonia mydas, in the Greater Caribbean. Ecol. Appl. 15, 304–314 (2005).
Naro-Maciel, E. et al. The interplay of homing and dispersal in green turtles: A focus on the southwestern atlantic. J. Hered. 103, 792–805 (2012).
Monzón-Argüello, C. et al. Evidence from genetic and Lagrangian drifter data for transatlantic transport of small juvenile green turtles. J. Biogeogr. 37, 1752–1766 (2010).
Luke, K., Horrocks, J. A., LeRoux, R. A. & Dutton, P. H. Origins of green turtle (Chelonia mydas) feeding aggregations around Barbados, West Indies. Mar. Biol. 144, 799–805 (2004).
Bass, A. L., Epperly, S. P. & Braun-McNeill, J. Green turtle (Chelonia mydas) foraging and nesting aggregations in the Caribbean and Atlantic: Impact of currents and behavior on dispersal. J. Hered. 97, 346–354 (2006).
Lahanas, P. N. et al. Genetic composition of a green turtle (Chelonia mydas) feeding ground population: Evidence for multiple origins. Mar. Biol. 130, 345–352 (1998).
Foley, A. M. et al. Characteristics of a green turtle (Chelonia mydas) assemblage in northwestern Florida determined during a hypothermic stunning event. Gulf Mex. Sci. 25, 131–143 (2007).
Bass, A. L., Lagueux, C. J. & Bowen, B. W. Origin of green turtles, Chelonia mydas, at ‘Sleeping Rocks’ off the Northeast coast of Nicaragua. Copeia 1998, 1064 (1998).
Bass, A. L. & Witzell, W. N. Demographic composition of immature green turtles (Chelonia mydas) from the East Central Florida Coast: Evidence from mtDNA markers. Herpetologica 56, 357–367 (2000).
Bjorndal, K. A., Parsons, J., Mustin, W. & Bolten, A. B. Threshold to maturity in a long-lived reptile: Interactions of age, size, and growth. Mar. Biol. 160, 607–616 (2013).
Perrault, J. R. et al. Maternal health status correlates with nest success of leatherback sea turtles (Dermochelys coriacea) from Florida. PLoS ONE 7, e31841 (2012).
Montero, N. et al. Warmer and wetter conditions will reduce offspring production of hawksbill turtles in Brazil under climate change. PLoS ONE 13, 1–16 (2018).
Shamblin, B. M. et al. Geographic patterns of genetic variation in a broadly distributed marine vertebrate: New insights into loggerhead turtle stock structure from expanded mitochondrial DNA sequences. PLoS ONE 9, 85956 (2014).
Anderson, J. D., Shaver, D. J. & Karel, W. J. Genetic Diversity and Natal Origins of Green Turtles (Chelonia mydas) in the Western Gulf of Mexico. J. Herpetol. 47, 251–257 (2013).
Acknowledgements
We are grateful to students, volunteers, and staff of the UCF Marine Turtle Research Group for collecting samples over the years, making this study possible. We thank Drs. D. Jenkins and S. Ceriani for thoughtful comments and suggestions during the experimental design phase, Dr. S. Ceriani for providing nest numbers laid in each Florida management unit used in the MSAs, and Drs. K. Phillips and B. Bolker for constructive feedback and helping to review the manuscript. We deeply appreciate Dr. L. "Doc" Ehrhart for decades of dedication, hard work, mentoring and, most importantly, for always having a dad joke handy when we needed it the most; he will be missed. This work was partially funded through grant #19-021R award by the Florida Sea Turtle License Plate Program, which is funded by the proceeds of the Florida Sea Turtle License Plate. G.D.S is a grantee of the ‘Ciência Sem Fronteiras’ scholarship program funded by CAPES (99999.013761/2013-07). This study was carried out in compliance with the ARRIVE guidelines.
Author information
Authors and Affiliations
Contributions
G.D.S., E.A.H., and K.L.M. designed the study and obtained funding, G.D.S. and M.R. contributed with field work and conducted DNA extractions and PCR amplifications, G.D.S. conducted data quality assessment and prepared figures and tables, G.D.S., E.A.H., and P.F.Q.-A. conducted statistical analyses, P.F.Q.-A. and G.D.S. modified the mixed stock model, K.L.M. supervised research and obtained permits and institutional animal welfare protocols. G.D.S. wrote the original draft, and all authors reviewed and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stahelin, G.D., Hoffman, E.A., Quintana-Ascencio, P.F. et al. Incorporating distance metrics and temporal trends to refine mixed stock analysis. Sci Rep 12, 20569 (2022). https://doi.org/10.1038/s41598-022-24279-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24279-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.