Abstract
Deep neural networks (DNNs) have shown success in image classification, with high accuracy in recognition of everyday objects. Performance of DNNs has traditionally been measured assuming human accuracy is perfect. In specific problem domains, however, human accuracy is less than perfect and a comparison between humans and machine learning (ML) models can be performed. In recognising everyday objects, humans have the advantage of a lifetime of experience, whereas DNN models are trained only with a limited image dataset. We have tried to compare performance of human learners and two DNN models on an image dataset which is novel to both, i.e. histological images. We thus aim to eliminate the advantage of prior experience that humans have over DNN models in image classification. Ten classes of tissues were randomly selected from the undergraduate first year histology curriculum of a Medical School in North India. Two machine learning (ML) models were developed based on the VGG16 (VML) and Inception V2 (IML) DNNs, using transfer learning, to produce a 10-class classifier. One thousand (1000) images belonging to the ten classes (i.e. 100 images from each class) were split into training (700) and validation (300) sets. After training, the VML and IML model achieved 85.67 and 89% accuracy on the validation set, respectively. The training set was also circulated to medical students (MS) of the college for a week. An online quiz, consisting of a random selection of 100 images from the validation set, was conducted on students (after obtaining informed consent) who volunteered for the study. 66 students participated in the quiz, providing 6557 responses. In addition, we prepared a set of 10 images which belonged to different classes of tissue, not present in training set (i.e. out of training scope or OTS images). A second quiz was conducted on medical students with OTS images, and the ML models were also run on these OTS images. The overall accuracy of MS in the first quiz was 55.14%. The two ML models were also run on the first quiz questionnaire, producing accuracy between 91 and 93%. The ML models scored more than 80% of medical students. Analysis of confusion matrices of both ML models and all medical students showed dissimilar error profiles. However, when comparing the subset of students who achieved similar accuracy as the ML models, the error profile was also similar. Recognition of ‘stomach’ proved difficult for both humans and ML models. In 04 images in the first quiz set, both VML model and medical students produced highly equivocal responses. Within these images, a pattern of bias was uncovered–the tendency of medical students to misclassify ‘liver’ tissue. The ‘stomach’ class proved most difficult for both MS and VML, producing 34.84% of all errors of MS, and 41.17% of all errors of VML model; however, the IML model committed most errors in recognising the ‘skin’ class (27.5% of all errors). Analysis of the convolution layers of the DNN outlined features in the original image which might have led to misclassification by the VML model. In OTS images, however, the medical students produced better overall score than both ML models, i.e. they successfully recognised patterns of similarity between tissues and could generalise their training to a novel dataset. Our findings suggest that within the scope of training, ML models perform better than 80% medical students with a distinct error profile. However, students who have reached accuracy close to the ML models, tend to replicate the error profile as that of the ML models. This suggests a degree of similarity between how machines and humans extract features from an image. If asked to recognise images outside the scope of training, humans perform better at recognising patterns and likeness between tissues. This suggests that ‘training’ is not the same as ‘learning’, and humans can extend their pattern-based learning to different domains outside of the training set.
Similar content being viewed by others
Introduction
Deep Neural Networks (DNN) have emerged as capable models for visual recognition; in fact, some have suggested that they are in-silico models of the human visual system1. The success of DNNs in recognition of everyday objects2 has raised speculations that activations in the intermediate layers of DNNs reflect the neural activity in visual cortex3. Such conclusions have often been drawn from observing DNN models that closely match human performance in recognition of everyday objects (cars, books, plants, animals, fruits etc.), i.e. those objects that the non-expert human is likely to encounter in his day-to-day life4. In some specialised domains, DNN models match or outperform humans: for example, there exist DNN models trained to perform a binary classification—‘tuberculosis’ or ‘healthy’—from chest radiographs5. However, such specialised models are of limited scope and do not generalise to the larger problem of visual recognition of random objects.
Since last decade, several DNNs have been developed which have been trained with huge image datasets, such as ImageNet6, and can recognise a large class of everyday objects, i.e. they can function as generalised object identifiers7. However, such DNNs are not free from errors8, and such errors offer a window into the inner mechanisms of the model. This presents an opportunity to compare DNN models with humans. Research on differences between machine and human perception has been carried out on macroscopic, everyday objects (both with normal and low intensity signals)9, gestures and motions10, and specially prepared image datasets for ease of comparison11. In some of these test conditions, such as noisy and distorted images, humans perform significantly better12. However, any such comparison is bound to suffer from a degree of bias, because human brains are trained for visual recognition since birth – and typically, recognition of everyday objects/symbols by a human is instantaneous, requiring almost no cognitive action. Machine learning models, however advanced, are trained on finite datasets that can never match the breadth and depth of the human visual experience. Thus, they are subject to several shortcomings: for example, a special class of images which have been subtly modified from the original, and which the human can discern quite easily, can often produce a drastically wrong result from a well trained DNN model. Such ‘adversarial’ images are increasingly becoming relevant in diverse domains such as cyber security and healthcare13. In addition, DNN models may classify a completely random array of pixels as a real-life object14, which raises speculations whether DNNs ‘perceive’ objects in the conventional sense of the word.
Pure perception, unbiased by prior knowledge, has been defined by several philosophic schools. In the Nyaya-sutra, an early philosophic text from India, the author Gautama mentions: “Perception is a cognition which arises from the contact of the sense organ and object and is not impregnated by words, is unerring, and well-ascertained” 15. Implicit in such a definition is that there should be no prior memory (‘word’) of the perceived object, in the ‘agent’ who perceives. The agent, in this case, might be a human or a machine learning (ML) model. Thus, any comparison between human and machine learning is bound to be affected by the bias of antedate, i.e. the prior experience of humans in recognition of everyday objects. We felt that to compare the performance of humans and DNN, a novel dataset, which is new to both humans and machines, is required.
It is in this under-explored niche that histological image recognition finds its use. Typically, human beings are not trained to recognise tissue type from histological images by default; only a very select subset of humans (medical students, zoologists) will ever learn how to recognise tissue type from histological images. And this presents a window of opportunity for comparison of human and machine learning, because both the machine and the human have had no prior experience in classifying histological images.
Brief review of relevant literature
The earliest precursor of DNNs is the Neocognitron developed by Fukushima et al. in 1980, which was the first model to implement a ‘shift invariant’ algorithm, i.e. detection of features was unaffected by the location of the feature in the image16. The name ‘convolutional neural network’ (CNN) was proposed by Lecun et al., who used the convolution operation and constructed a simple network (LeNet) consisting of convolution-pooling-convolution-pooling-dense-dense layers, for handwritten digit recognition tasks17. However, image classification gained traction since publication of the ImageNet database, a large, multi-class collection of images, in 2009. The first influential CNN model, AlexNet, an 8-layered CNN, achieved 15.3% error-rate on the ImageNet database 18. After a few years, The Visual Geometry Group (VGG)19proposed several deep neural networks (11 to 19 layers deep) which achieved 7.3% error rate on the ImageNet database. In 2014, the Inception block architecture (a local unit with parallel branches of convolution) was introduced by Google Inc. in their models Inception V1 to V4, which reduced the error rate to 6.7%20. A slight improvement on the Inception model was proposed by Chollet in 2017, introducing depthwise convolution followed by pointwise convolution, termed as the Xception model21. The Xception model achieved 79% top-1 accuracy on the ImageNet database, and has been employed in several other problem domains such as finger vein recognition22 and plant disease classification23. The CNN paradigm was further improvised with the introduction of residual nets (skip connection between layers) in 2015 by He et al.24 In a further improvement, Tan et al., in 2019, developed the EfficientNet, which implemented a new scaling method for CNNs, achieving 88% top-1 accuracy in Imagenet25. A variant, the FixEfficientNet by Touvron et al., which corrects discrepancy between training and test images, achieved 98.7% top-5 accuracy on ImageNetV226. In early 2020s, there was another paradigm shift in image classification: Dosovitskiy et al. proposed implementing the transformer architecture, used in natural language processing tasks, in image classification27. Such Vision Transformers (ViTs) have now become state of the art in image classification, producing 90.94% top-1 accuracy on ImageNet database28.
Similar developments have taken place in feature extraction and enhancement of images. The extreme gradient boosting classifier (XGBoost), developed by Chen et al.29, was used on the Caltech-101 image dataset producing high accuracy (88.36%), outperforming most other major feature detectors30. Kumar et al. have demonstrated feature extraction from images has advanced sufficiently to retrieve images from large datasets based on content, with 99.53% precision31. Low contrast images, such as underwater photographs, have been demonstrated to be enhanced by the Contrast-Limited Adaptive Histogram Equalization (CLAHE) algorithm, which is a significant improvement over conventional histogram equalisation method32,33. Alternative approaches to image classification, such as the Shi-Tomasi corner detection algorithm34 along with the scale-invariant feature transform (SIFT)35, have produced 86.4% accuracy on the Caltech-101 image dataset36. It’s also been demonstrated that image analysis techniques can detect the device of origin (i.e. camera brand) with reasonable accuracy37. Such advances in image analysis techniques have complemented the ML models for successful image classification.
Neural networks are not limited to the domain of image analysis; they have also found application in signal processing, such as biometric identification through electrocardiogram (ECG)38,39 and analysis of electroencephalograms40, often in conjunction with other ML models such as support vector machines and k-nearest neighbours41. The generic nature of neural networks provides them with extensibility to suit diverse problem domains.
In the specific field of histology, the application of deep neural networks has been explored in recent studies, albeit in a fragmentary manner. A limited histology classifier was developed by Rujano-Balza et al., which classified into five basic types (epithelium, gland, connective, muscular and nervous tissue)42. The VGG16 model, the one we have used in the present study, has been previously used in classifying breast cancer histology images with 92.60% accuracy (albeit as a binary classifier: benign versus malignant)43. Ahmed et al. have used the VGG16 and InceptionV3 pretrained models to classify histological images from a mixed dataset (Kimia Path 24)44; however, their method was not targeted to identify specific tissues (the Kimia Path dataset is a mixed set of images from different tissues with different stains). We identified that there was a definite lacuna in literature regarding comparison of human versus ML performance in this specific domain. Thus we took the approach of multi-class tissue classification on a defined set of tissues, similar to the training imparted to medical students in first year of medical school.
Problem statement
We wanted to compare the performance of two kinds of ‘agents’ on a novel dataset. The agents are.
1. Machine learning model (ML)—two pretrained CNNs.
2. A group of medical students (MS).
We have compared the performance of MS and ML models on a validation set – composed of tissues within the scope of training; we have also compared their performance on a smaller set of images outside of the scope of training, i.e. a completely different set of tissues not included in training. A random sample of images used in the study (and included in the training set) is shown in Fig. 1.
Random sample of images used in the study: (a). Trachea, (b). Cerebrum, (c). Stomach, (d). Kidney, (e). Cerebrum, (f). Heart, (g). Stomach, (h). Skin, (i). Lungs, (j). Trachea (Composite figure generated with ImageMagick, version 7.1.0, https://imagemagick.org/index.php).
The objective of the present study was to (a) compare the accuracy of ML models and medical students in an image classification problem, on a dataset which is novel to both humans and ML models, (b) compare the pattern of errors of ML models and medical students and thus find similarities, if any. This would provide an insight whether there are similarities between the inner representation of visual information in humans and ML models.
Materials and methods
Ethics statement
Ethical clearance was obtained from Institutional Ethical Committee, Base Hospital and Army College of Medical Sciences, Delhi Cantonment, Delhi, India (No. IEC/01/2021/10). The study involved voluntary and anonymous participation by medical students in an online quiz; students were asked by the faculty of Dept of Pathology, Army College of Medical Sciences, to participate in the study on a purely voluntary basis. Informed consent was obtained from participants via electronic medium. The quiz was fully anonymised: no personal information, which could potentially reveal the identity of the candidates, was collected in the online quiz.
The study did not involve any kind of diagnostic/therapeutic modality on human subjects. A group of adult medical students were asked to voluntarily and anonymously participate in an online quiz, hosted by Army College of Medical Sciences, after adequate information to students. All relevant guidelines regarding participation of human subjects, including the Helsinki declaration, were followed.
Preparation of image dataset
Histological images from tissues were collected from the archives of a hospital in North India. An Olympus Magcam Microphotography system was used for acquiring the images. All images were acquired with a Dewinter 606 Trinocular Microscope, under the same condition of illumination and 10 × maginification. The images were acquired in 1280 × 960 pixels resolution and resized to 512 × 384 pixels with ImageMagick image processing software45. All histological slides were anonymised before image acquisition.
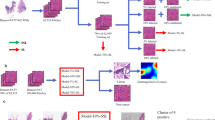
For selection of tissues for inclusion in the study, we made a list of tissues which are introduced to the medical students in first year curriculum, and randomly selected 10 classes from them. Images from the following ten classes of tissue were acquired: cerebellum, cerebrum , heart , kidney , liver , lung , pancreas , skin , stomach , trachea. One hundred (100) images from each class were acquired, for a total of one thousand (1000) images. The schematic diagram of the study is presented in Fig. 2.
Schematic diagram of the study (Figure generated with Libreoffice Draw, version 7.2.7, https://www.libreoffice.org/).
The image dataset was randomly split into two subsets (using the ‘shuf’ program of the Linux operating system)46:
I. A training set of 700 (seven hundred) images, consisting of 70 images from each class of tissue.
-
(a)
Within the training set, 20% images (140 images) were selected randomly (using the train_test_split function from the scikit-learn Python library, version 1.0.247) as test set, for concurrent testing during training of ML model.
II. A validation set of 300 (three hundred) images, consisting of 30 images from each class of tissue.
-
(a)
From the validation set, 100 images were marked for inclusion in the quiz set, i.e. for testing human participants; the quiz set contained 10 images randomly selected from each of the classes ‘cerebellum', ‘pancreas', ‘kidney', ‘trachea', ‘cerebrum', ‘lung', ‘heart', ‘skin', ‘liver' and ‘stomach'.
Construction of ML model
Two ML models were constructed using the DNN architecture. We used transfer learning in constructing the ML model, i.e. used the convolutional layers of a pretrained model for image analysis (i.e. to extract features of the image). Initially, we shortlisted two models, pre-trained VGG16 and ResNet50, because both of them are extensively trained with ImageNet database and frequently used in image recognition tasks. The output of the convolution-pooling layers from both models was flattened into a one dimensional array, and several fully connected and dropout layers were added to produce a final output layer of 10 classes. During initial testing, the modified ResNet50 model did not exceed 50% accuracy on the concurrent testing set, and thus it was not selected for the study. We selected VGG16 as our model, because in addition to its good performance on the testing set, it has been used previously in histological classification problems (Shallu43). In addition, VGG16 was used by a similar study by Dodge et al., which used everyday images (i.e. dog breeds) and their distorted versions, for human–machine comparison12.The architecture of the model is shown in Table 1. In joining the convolution – pool layers of VGG16 and the fully connected layers, the method published by Rosebrock48 and Brownlee49 was followed.
The input image was preprocessed with the OpenCV image processing software50, which consisted of resizing the color image to 128 × 96 pixels. The DNN model takes this numeric array as input; thus its input dimension is 128 × 96 × 3 (the third dimension corresponds to the three color channels, red, green and blue, at each pixel). After several convolution and max-pooling layers, the array is reshaped to a shape of 4 × 3 × 512. This layer is flattened to a one-dimensional array of size 6144. Several fully connected and dropout layers then reduce the array to a final size of 10. Figure 3 shows the architecture of the modified VGG16 model, represented graphically.
Graphical representation of the modified VGG16 model; the model takes a 128 × 96 color image as input (extreme left) and converts it to a 10 class output (extreme right); the dark blue block represents the flattened output of the convolution and max pooling layers of VGG16, which is followed by fully connected and dropout layers (Figure produced with the Visualkeras python library, version 0.0.2, https://github.com/paulgavrikov/visualkeras).
The model was constructed with the Keras deep learning library (version 2.8.0)51 in Python programming language, using the Google Colaboratory platform.
In addition, after rejection of ResNet model, we used another pretrained model (Inception V2) from the Keras applications library; we modified it in a similar manner as the VGG16 model, i.e. flattened its output layer and added a series of dense and dropout layers. The structure of the modified inception model is shown in Table 2.
Training of VGG 16 model
The model thus developed was trained with the training dataset over 30 epochs, with a batch size of 10 images. At the end of training, the model achieved 81.96% accuracy over the training test, and 77.14% accuracy in the concurrent test set. Accuracy and loss (error rate) of the model over epochs of training are shown in Fig. 4.
Accuracy and loss (error rate) of the VML over 30 epochs of training (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Evaluation of VGG16 ML (VML) model
The VGG16 model (VML) was evaluated on the validation set (n = 300), which yielded 85.67% accuracy. The confusion matrix on the validation set is shown in Table 3:
The VGG16 model (VML) classified 257 of the 300 images in the validation set correctly (85.67%). The commonest error by the VGG16 model was (a) misclassifying stomach as liver (08 images) and (b) misclassifying kidney as liver (08 cases) and (c) misclassifying stomach as heart (04). The best accuracy was recorded in recognising lung (30/30, 100%) and the worst in stomach (15/30, 50%).
Figure 5 shows examples of correct classification by the VML. The VML outputs 10 numbers, corresponding to the probabilities of the image belonging to the respective class of tissue. In Fig. 5a, the classification is unequivocal, i.e. there is a very high probability (close to 1) of the tissue being ‘skin’. In Fig. 5b, the VML has generated various probabilities for different classes of tissue, i.e. the output is equivocal, and the class with the highest probability (‘kidney’) has been chosen as the correct answer.
Examples of classification by VML: (a) Skin, correctly identified by the VML; the artifact in the section has not interfered with correct classification by the VML; (b) Kidney tissue as classified by the VML; the model generates 10 numbers as output, corresponding to the probability of 10 classes of tissue. In this case, 'liver' is a close second (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Training of inception V2 model (IML)
The inception V2 model was trained with the same training dataset as the VGG16 model with same hyperparameters (30 epochs, with a batch size of 10 images). The model achieved 89% accuracy on the concurrent testing set (Fig. 6).
Accuracy and loss over 30 epochs of training the Inception model; this model showed a much sharper drop in error rate (loss) during the early epochs, than the VML model (Fig. 4) (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Evaluation of Inception model
The Inception model (IML) achieved 90.33% (271 out of 300) accuracy on the validation set. In contrast to the VGG16 model, the Inception model made maximum number of errors in recognising the ‘skin’ class (8 errors out of 29, 27.5%), followed by ‘stomach’ class (7 errors). (Table 4) Examples of classification by the IML model are shown in Figs. 19, 21 and 23.
Training of medical students (MS)
The medical students (MS) of who have just completed first year in a Medical School in North India were asked to volunteer for the study. We circulated the Participation Information Sheet by electronic messaging through the class representative. It was explained to the students that the very act of submitting the electronic questionnaire will be taken as consent for participation in the study. The purpose and objective of the study was explained to the class, and that anonymised data from the study will be published. Informed consent was obtained from participants via electronic medium. The students were trained with basic histological tissue identification for a period of 02 months before the study, as part of their curriculum in 1st year MBBS course. The training of medical students in histology was imparted in a heuristic manner as is conventional in histology: specific identification criteria were taught to the students—i.e. ‘liver’ was to be identified by its lobular structure and kidney by glomeruli etc. In addition, the complete set of 700 training images were circulated to medical students one week prior to the quiz.
In our present medical curriculum, first year medical students are introduced to human histology. Majority of these students have never seen histological images of human tissues before, and are thus a naive population. Their performance in recognising histological images is very similar to a naive, untrained ML model. These students are ideal candidates for training with a set of histological images and assess the resulting efficacy, for direct comparison with a similarly trained ML model. By the time they complete second year of medical school, they have already been well trained with histological images as part of the pathology curriculum, and thus cannot be compared with an ML model.
Preparation of questionnaire
The ‘quiz’ set (IIa) of images (100 images) were incorporated into an online quiz format (built with Microsoft Office Forms); a set of 100 questions were formulated, with a choice of 10 options each, corresponding to the ten classes of tissue. Answering all questions was not mandatory (and all such ‘blank’ or ‘other’ answers were excluded from final analysis); no time limit was set on the questionnaire. (Link to the questionnaire is provided in the ‘Data availability statement’ section).
Conduct of survey
The quiz questionnaire was shared online with medical students. The online survey was conducted throughout a week. Strict anonymisation was maintained in data collection; no personal information was collected from the survey. However, students were strictly instructed to respond to the quiz only once per person, i.e. no duplication in responses was allowed. At the end of the week, sixty-six (66) responses were aggregated and tabulated.
Preparation of out-of-training-scope (OTS) image set
As adversarial examples, meant to test the pattern recognition capabilities of ML and MS, 10 images from tissues which were not part of the training data were separately selected from archives of the hospital. The 10 images were as follows (Table 5):
The OTS images were selected to highlight the differences between ML and MS. The ‘intestine’ class was included because of its similarity to the ‘stomach’ class, and also that medical students are likely to have seen images of intestine before, while the ML model has never been trained with ‘intestine’ class. The ‘cervix’ class was chosen because of its similarity with ‘skin’, and ‘salivary gland’ because of its similarity to ‘pancreas’. The parathyroid and endometrium was unlike any tissue the students or ML models would have encountered during training.
Evaluation of ML and MS on OTS images
Both ML models were run on the OTS images and the predictions were recorded. A second online quiz was conducted on the medical students with the OTS image set. 44 responses were recorded.
Informed consent
The study involved voluntary and anonymous participation by medical students in an online quiz; students were asked by the faculty of Dept of Pathology, Army College of Medical Sciences, to participate in the study on a purely voluntary basis. Participant information sheet (PIS) was circulated to the students; informed consent was obtained from participants via electronic medium. We circulated the Participation Information Sheet by electronic messaging through the class representative. It was explained to the students that the very act of submitting the electronic questionnaire will be taken as consent for participation in the study. The purpose and objective of the study was explained to the class, and that anonymised data from the study will be published.
Results
Performance of VML model on quiz questionnaire
The VGG16 ML model achieved 91% accuracy in the quiz (Table 6). The most common error encountered was misclassifying kidney tissue as liver. The model showed poorest performance in recognising stomach: misclassifying stomach tissue as heart (02 images), liver (01 image) and pancreas (01 image).
Performance of IML model on quiz questionnaire
The IML model produced 93% accuracy on the quiz questionnaire (Table 7). In contrast to the VML model, the commonest error of the IML model is recognising ‘skin’ (03 errors), which was variably mistaken as ‘kidney’ (02 errors) or ‘stomach’ (01 error).
Performance of medical students on quiz questionnaire
The combined performance of 66 medical students on the quiz questionnaire (a total of 6557 responses, excluding blank / ‘other’ answers) is shown in Table 8. The overall accuracy of the medical students (MS) was 55.14%.
Analysing by responses from individual students, accuracy of students ranged from 4 to 100%. 13 medical students out of 66 (19.67%) matched or exceeded the accuracy of the VML model, i.e. the VML model was at 80th percentile if compared with medical students (Fig. 7). Only 08 medical students (12.12%) scored better than the IML model.
Accuracy of 66 medical students in ascending order (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
A histogram of the accuracy of medical students shows concentration of scores at two extremes of accuracy (Fig. 8).
Histogram of accuracy of 66 medical students in the quiz questionnaire; the accuracy of VML and IML are shown for comparison (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Comparison of ML models and medical students on questionnaire
Analysis of responses, student-by-student, shows variable agreement with the VML model (Cohen’s kappa ranging from − 0.06 to + 0.9). Negative kappa values were seen in Participants 58, 38, 39, 40, 43, 45 – i.e. the same students who had achieved lowest test scores (Fig. 9).
Plot of Cohen’s kappa for each student (n = 66), as a measure of agreement between students and VML model (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
A tissue-wise comparison between MS (aggregate of all responses) and both ML models (VML & IML) showed the classification by ML models to be significantly different from MS; results of tissue-wise Chi-square Goodness of Fit are shown (Table 9):
Image-by-image comparison of equivocal responses by MS & VML
Figure 10 shows the most frequent answer among all medical students, and the most probable prediction from VML model, for each image, as a clustered heatmap.
Clustered heatmap showing commonest responses by MS, and predictions with highest probability from VML, on each individual image of the quiz set (n = 100); 90% match is noted (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The comparison between 2nd commonest answers from MS, and predictions with 2nd highest probability from VML, image-by-image, is shown in Fig. 11.
The second commonest answer of the medical students, and the prediction from VML model with second highest probability—matched only in 15 images of the quiz questionnaire (n = 100); in 14 of these images, the first prediction also matched (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
A plot of top 3 commonest answers, per question basis, from both MS and VML, shows a diffuse scatter. No clustering is noted between the top 3 answers/probabilities of MS and VML, on an image-by-image basis (Fig. 12).
Plot of three commonest responses from MS and VML. Each dot represents a question from the quiz. The x axis represents the commonest response, y axis – the second commonest, and z axis – the third commonest response, from medical students on that question (in blue). For VML model (in green), the x axis is the prediction with highest probability, y axis – second highest and z-axis, third highest probability. The numbers 0 to 9 on each axis represent the 10 categories of tissue: 'cerebellum', 'cerebrum', 'heart', 'kidney', 'liver', 'lung', 'pancreas', 'skin', 'stomach', & 'trachea' . The quiz consisted of 100 questions (n = 100) (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The IML model was decisive in its predictions in majority of the cases: only 02 questions out of 100 in the quiz set were answered equivocally, i.e. where the prediction with the 2nd highest probability was more than half of the first. Thus the IML model was not comparable to MS regarding equivocal responses.
Images that were difficult to classify, for both MS and VML
We found only 04 (four) images where both MS and VML have produced a grossly mixed response, i.e. the frequency (probability) of the second choice was more than half of that of the first choice. The second choices of MS and VML were different in all such images (Fig. 14). Three of these images belong to the class ‘liver’. While both MS and VML model have generated correct classification with the greatest probability, it is interesting to note that the 2nd most frequent answer from MS was always ‘pancreas’, and the 2nd highest probability from VML model was always ‘kidney’. ‘Stomach’ is the 3rd most probable prediction in all three images, by both MS and VML (Fig. 13).
The four images in the quiz set where both medical students and VML model have produced mixed responses, with the 2nd commonest response having more than half the probability/ frequency of the commonest response. (a) stomach, (b), (c) & (d) liver. (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Comparison of MS on quiz questionnaire and VML/IML on full validation set
The ten commonest errors in classification by aggregate of MS, as compared to VML model (the results of the VML model on the full validation set of 300 images, not just the quiz images) & the IML model (on the full validation set), are shown in Table 10:
The ‘stomach’ class accounted for the highest number of errors in both VML and MS, producing 34.84% of all errors by MS and 41.17% of all errors by VML model. The commonest error by medical students was misclassifying stomach as pancreas (160 errors), liver as pancreas (112 errors), and cerebellum as cerebrum (101 errors) (Fig. 14).
Error patterns of MS, VML & IML: (a) commonest errors of the medical students (MS) over the quiz set (n = 6557); (b) commonest errors of VML on the validation dataset (n = 300); (c) commonest errors of the IML on validation dataset (n = 300); a label such as ‘liver/ kidney’ indicates that ‘liver’ was mistaken for ‘kidney’ (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The commonest error of the VML model on full validation set were misclassifying kidney as liver (08 errors), misclassifying stomach as liver (08) & misclassifying stomach as heart (04). There is no overlap between the commonest errors of MS and VML model (Fig. 14). The pattern of errors of the IML model is distinct from both MS & VML; ‘skin’ is the commonest misidentified class, followed by liver and heart (Fig. 14).
Comparison with students with scores close to ML models
The overall accuracy of MS was much lower than the VML model/IML model, and thus their confusion matrices were not directly comparable. We selected 05 students whose accuracy in quiz was in the range 0.90–0.92, i.e. close to that of the VML model. We constructed their aggregated error matrix (Table 11).
We then compared this confusion matrix with the error matrix of the VML and IML model over the entire validation set (Table 3). A Kolmogorov–Smirnov test showed K-S statistic 0.1, p-value = 0.702, suggesting similar error profiles between the VML/IML and this subset of medical students.
Results of OTS images
The results of MS and VML on OTS images (10 images) are as follows. A ‘likeness score’ has been assigned based on resemblance of original and predicted tissue, i.e. if ‘salivary gland’ is predicted to be ‘pancreas’, a likeness score of 01 is allotted (Table 12).
The same set of images was shown to the MS and their responses recorded. As shown in the table, the VML & IML model have clearly overfitted on the class ‘lung’. This is concordant with the perfect accuracy (100%) of both VML & IML in recognising lung tissue in the validation set. Unlike the VML model, majority of medical students have been able to place the images in the category which it resembles histologically, i.e. they have consistently classified intestine as ‘stomach’ (possibly, they could recognise intestine, but selected ‘stomach’ because the options were limited to only ten classes of the training dataset). In recognising ‘parathyroid’ and ‘endometrium’, MS, VML & IML model have produced random results (Fig. 15); this is expected – as the VML/IML model was never trained on these tissues. Also, the medical students were not trained on these tissues (parathyroid, endometrium).
Parathyroid tissue predicted by VML & IML to be ‘lung'; the opinion of medical students is split between several classes (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Discussion
The emergence of DNNs has rekindled academic interest in the true nature of perception and working of the mind. Qualities that were erstwhile known to be decidedly biological, such as visual recognition of objects, have been successfully reproduced in DNN models. Even the most abstract of faculties, mathematics and symbolic logic, have been implemented with DNNs with some degree of success52. However, a direct comparison of DNN with human visual recognition is difficult because of several reasons. Humans, although well trained by a lifetime of experiences, are prone to the occasional error in visual recognition (such as optical illusions, wrong interpretation of color, even misrecognition of faces53). Adversarial images, which can confound DNN models, have also been shown to affect humans when limited by time for decision-making54.
In early studies on DNNs, the performance of humans was assumed to be 100% accurate, i.e. human visual recognition was the gold standard against which DNN models were compared. However, it was soon realised that in specialised domains (such as recognising dog breeds), humans were just as naive as machines, and DNN models may outperform humans55. But even in such studies, the base class ‘dog’ is not outside the purview of human experience, although the subclasses (breeds) might be unknown to the human subjects. Thus, there still remains a bias in favor of humans.
Funke et al., in their guideline, describe the control variables when comparing human and ML performance on image recognition56. One such control is ‘aligning experimental conditions for both systems’, i.e. the matching operation. They mention that ‘the human brain profits from lifelong experience, whereas a machine algorithm is usually limited to learning from specific stimuli of a particular task and setting.’ The same point was highlighted by Cowley et al. in a recent paper on designing a framework for human–machine learning comparison. They observed that while the ML model can be trained only with a limited set of data, the same does not apply to humans and it was not possible “ … to implement a one-shot learning task in human participants using natural images and categories that humans already have experience with” 57. We have tried to remove the bias of experience, by studying an image dataset which is novel to both human and machines (i.e. histological images).
However, it is to be emphasised that the training period differed between humans and machines, in our study. This is due to the differences in machine and human learning: we have selected medical students who had already been taught histology as part of their curriculum in first year of medical school. This is because humans need a minimum period of training to gain proficiency, and to develop pattern recognition skills which they can generalise to novel problems. An ML model is much faster to train (typically, within hours or days); humans cannot be expected to learn anything within such a short period.
Pattern of errors in MS & VML/ IML model
Overall, the pattern of errors of medical students and VML/IML model is distinctly different, as seen by the overall kappa and chi-squared Goodness of Fit tests (Table 9). In the quiz questionnaire, there were only 04 images which generated mixed response both from MS and VML. However, the pattern of the responses (i.e. 2nd and 3rd common response) was different in MS and VML. Whereas the commonest error by MS was stomach/ pancreas (i.e. classifying stomach as pancreas) and kidney/pancreas, the commonest errors of the VML model was kidney/liver and stomach/ liver. This suggests ‘overfitting’ on pancreas by MS, and on liver by VML model.
Interestingly, the images of ‘stomach’ class were obtained from gastric biopsies, and were thus missing muscularis propria. Thus, the ‘stomach’ class has assisted us to uncover hidden bias in both MS and VML. The medical students tend to mistake the stomach for pancreas, whereas the VML model frequently mistook the stomach for liver.
The IML model, however, is much more certain in its responses (only 2% equivocal responses in the quiz set). It shows a bias for the ‘kidney’ class, and has frequently mistaken skin and liver for kidney. This pattern is also different from medical students. Because of its higher accuracy and greater certainty than VML, we felt that the IML model was less suitable for comparison with medical students.
Visualising deeper layers of the DNN in misclassified images
Following is a serial visualisation of convolutional layers of the VML, which is processing an image of trachea, and misclassifies it as cerebellum. The MS group have however, correctly recognised this image as trachea with a clear majority (Figs. 16 and 17).
An image of trachea recognised correctly by majority of MS, misclassified by VML and correctly classified by IML (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The VML model processing the image from 'trachea' class (Fig. 16), eventually misclassifying it as ‘cerebellum’. The inner layers (convolution layers only) are represented here. The thick band of cartilaginous tissue is the prominent feature of this image, and has persisted till the last convolution layers. (For visualisation purposes, a psuedocolor scheme has been used to render the deeper layers, and only the first few slices of initial convolution layers are shown) (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
We compared this with an image of ‘cerebellum’ class that was correctly classified by the VML. The similarities in the convolutional layers are evident. It is possible that the cartilaginous band in trachea led to the misclassification of the previous image as cerebellum, due to similarities with the granular layer of cerebellum (Figs. 18 and 19).
An image of cerebellum correctly classified by majority of MS, VML and IML (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Image of 'cerebellum' class correctly classified by the VML; there is similarity of the initial first 10 slices of the first convolution layer, with the previous figure (Fig. 18). The granular layer has persisted till the last convolution layers. (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
This is in stark contrast to an image from ‘liver’ class, which was recognised by most of the students, and also by VML model with a mixed response (Fig. 20). The IML model, however, unequivocally classifies this as ‘liver’.
Image from ‘liver’ class producing mixed response from MS & VML but unequivocal response from IML (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Deeper layers of the VML reveal specific signatures of this image which do not persist till the innermost layers (Fig. 21).
Convolution layers of the image in previous figure (Fig. 20) within the VML. No specific identifying feature has persisted till the last few convolution layers (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The ‘stomach’ tissue, which has turned out to be the most confounding in both humans and VML model, was mistaken for all other classes of tissue. Figure 22 shows stomach tissue mislabeled as liver by the VML model. The first few convolutional layers show the artifactual blank space which has given the gastric mucosa a vague lobular appearance. (Fig. 23).
Stomach mucosa producing mixed response from MS, wrongly identified by VML and correctly identified by IML (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The artifactual linear blank space in the previous figure (Fig. 22) has prominently featured in the first few convolutional layers of VML model (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
Comparison of errors of MS and ML models
An interesting finding in the study was that students who have scores close to the VML/ IML model (i.e. between 0.9–0.92), produce similar error profile of the VML/IML model (when the performance of VML/IML model over entire validation set is considered). A Kolmogorov–Smirnov test failed to reject the null hypothesis (p = 0.702)58. However, when compared with all medical students, the pattern of errors was distinctly different. (Fig. 24).
Error matrices of VML, MS and IML: (a) Confusion matrix of VML over the entire validation set (n = 300); the dark spots along the diagonal represent correct answers; (b) Aggregated confusion matrix of medical students over the quiz images (n = 6557); dark spots are noted outside the diagonal line, representing wrong answers; (c) Confusion matrix of IML model on validation set (n = 300); the IML is more accurate, and shows less equivocal predictions, than the VML model (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
In a study by Dodge et al. on image dataset of different dog breeds, not only did humans outperform machines on serially distorted images, but the pattern of errors was also significantly different between humans and machines;12 the authors went on to suggest that the inner representation of images vary between humans and machines. However, the accuracy of the DNN model in their study was much lower than humans. In the present study, very few students have reached accuracy close to the machine (Fig. 25); between these few students and VML/ IML model, the error profile was similar.
Confusion matrix of all 66 students over the quiz set; the overall error profile is very dissimilar to ML models; only the best performing students produced a similar error profile as that of the VML model (Figure generated by Matplotlib python library, version 3.3.4, https://matplotlib.org/).
The IML model, however, in addition to being more accurate than both MS & VML, also shows a distinct error profile (with a bias for the ‘kidney’ class). The IML model seems to be more ‘certain’ in its decision than VML model and aggregate of medical students. Whereas the aggregate of medical students provided equivocal answers to 12 questions (12%) of the quiz set, and the VML produced equivocal predictions in 20 questions (20%), the IML generated an equivocal prediction in only 02 questions (2%). (By ‘equivocal’, we mean where the second commonest answer/prediction was ≥ 50% of the commonest answer/prediction–in frequency).
Likeness score of MS, VML & IML model in OTS images
In the OTS image set, we created a metric ‘likeness score’ based on histologic similarity of actual and predicted classification: if ‘cervix’ was recognised as ‘skin’, a likeness score of 01 was awarded (owing to presence of squamous epithelium in both of the tissues). The medical students (total likeness score = 6) outperformed the VML/IML models (total likeness score = 2). This was due to overfitting of the ML models on the ‘lung’ class (07 & 08 of the 10 images in the OTS set was classified by VML and IML model as ‘lung’, respectively), as well as indicating the higher efficacy of humans in pattern recognition from OTS images (i.e. finding similarities of architecture between ‘salivary gland’ and ‘pancreas’, both of which are exocrine glands).
The failure of the VML in recognising ‘likeness’ between tissues hints at the same problem faced by Funke et al. while trying to teach DNN models the concept of ‘closed’ and ‘open’ shapes;34 they realised that even after intensive training, the DNN model did not learn the concept of a closed shape. In our study, the overfitting on ‘lung’ class suggests that while the VML/IML model can recognise lung and pancreas with reasonable accuracy, it has not learnt the concept of a glandular structure, as evident by its misclassification of ‘salivary gland’ as ‘lung’.
Comparison with previous studies
Several efforts have been made in the recent past, to compare human and machine learning in the field of image recognition. The design of these studies varies in their problem space and models used, and are thus not directly comparable to the present study. (Table 13) However, our conclusions are similar to the studies by Kuhl 2020, Dodge 2017 and Fleuret 2011: that humans generalise to a larger problem space much faster than ML models, as evident by performance of medical students on the OTS image data.
Limitations
The study was limited by the quality of tissue preservation: in particular, the images from ‘liver’ and ‘kidney’ class were retrieved from autopsy specimens and some distortion of tissue architecture was present in few of the images. The images from ‘stomach’ class were from mucosal biopsies, resulting in loss of orientation of tissue. It must be mentioned that the ‘stomach’ class of tissue has, inadvertently, uncovered hidden bias in both MS and VML model, as mentioned in results.
Again, the medical students represent variable amounts of prior knowledge of histology, as is expected in any group humans with diverse academic aptitudes. This has reflected in the variation in accuracy of medical students. It is difficult to correctly match the exact amount of ‘training’ between humans and machines: we have selected medical students with some prior training in histology, i.e. trained for 2 months in histology as part of their curriculum in first year of medical school. This is because an initial, informal assessment on completely naive students had shown that without a period of training of at least a month, human students did not achieve > 10% accuracy in recognising 10 different classes of tissue. Certainly, the time taken by the VML/IML model to train on these tissues (2 h) is grossly insufficient for human students.
The quiz was conducted without any time limits, and answering each question was not mandatory (unanswered questions were excluded from final analysis). With the didactic experience of three of the authors, the psychology of students when facing a quiz was taken into consideration. We thus felt that making questions mandatory, or putting a time limit, would force the students to cause mistakes. It was decided not to implement artificial restrictions on human students. It is at this point that ML models significantly differ from medical students: an ML model, given an image, will always produce an answer; it cannot ‘pass’ questions. This is a fundamental limitation in comparing human and machine learning. However, we decided to mitigate this limitation by excluding unanswered questions (from medical students). In the course of analysis, especially when calculating Cohen’s kappa, unanswered questions by students were placed into the ‘other’ category and counted as errors. It must also be noted that even without making an answer mandatory, 6557 responses were received from medical students out of a possible 6600 (99.34%).
Conclusion
The present study compares the performance of two deep learning models (a modified VGG16 model and a modified Inception model) with a group of human medical students, on an image dataset which is novel to both: the deep learning models and humans have had no prior exposure to histological images. Our findings suggest that within the scope of training, the deep learning models perform better than 80% medical students. The medical students (MS) and VGG16 model (VML) faced similar difficulties in classification, as evident by the fact that ‘stomach’ was most difficult to classify by both of them, although the error profile–what they mistake it for–was different between MS and VML; in contrast, the Inception model (IML) was most frequently confounded by the ‘skin’ class. However, the students who have reached accuracies close to the VML/IML, also tend to replicate the same pattern of errors in image recognition as that of the ML models. This suggests a degree of similarity between how an ML model and humans extract features from an image when their accuracies are similar.
If asked to classify images outside the scope of training, humans perform better at recognising patterns and likeness between tissues. We suggest that ‘training’ (in the context of machine learning) is not the same as ‘learning’, and humans can extend their pattern-based learning to different domains outside of the training set.
Link to training notebook (annotated source code) of the DNN models, the VGG16 and Inception models (in H5 format) is provided in Data Availability section.
Future directions
The present study offers an insight into the inner representation of visual information, of human and machine learners, on a novel dataset. The study can be extended to any direction, particularly with state of-the-art-image classification models such as Xception and Vision Transformers. It would also be interesting to reproduce this study with altered or fragmented images, to assess performance of both the learners in recognising local features from an image. Such studies will bring the differences between humans and machine learning models, in the context of image recognition, in sharper focus.
Data availability
Quiz questionnaire with images: https://forms.office.com/Pages/ResponsePage.aspx?id=DQSIkWdsW0yxEjajBLZtrQAAAAAAAAAAAAe__YDJd8ZUMUJDU1JOUjJBUzBISDk5UDJaRkIwUjhITi4u. OTS questionnaire with images: https://forms.office.com/Pages/ResponsePage.aspx?id=DQSIkWdsW0yxEjajBLZtrQAAAAAAAAAAAAe__YDJd8ZUMU9IVVNWRUtRU0I2MEMyMUFLTExZVkhXTC4u. Annotated source code of the DNN models, the responses from students on quiz and OTS images are uploaded at: https://github.com/cmacus/histo_ml_human. Any other data pertaining to the study will be made available on request.
References
Cichy, R. et al. Comparison of deep neural networks to spatio-temporal cortical dynamics of human visual object recognition reveals hierarchical correspondence. Sci. Rep. 6, 27755 (2016).
Redmon, J., Divvala S., Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. Preprint at https://arxiv.org/abs/1506.02640 (2016).
Güçlü, U. et al. Deep neural networks reveal a gradient in the complexity of neural representations across the ventral stream. J. Neurosci. 35(27), 10005–10014 (2015).
Yamins, D. L. K. et al. Predicting higher visual cortex neural responses. Proc. Natl. Acad. Sci. 111(23), 8619–8624 (2014).
Lakhani, P. & Sundaram, B. Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284(2), 574–582 (2017).
Deng J., Dong W., Socher R., Li L.J., Li K. and Fei-Fei L. ImageNet: A large-scale hierarchical image database. IEEE Computer Vision Pattern Recognit., 248–255 (2009).
Alom M.A., et al. The History Began from AlexNet: A comprehensive survey on deep learning approaches. Preprint at https://arxiv.org/abs/1803.01164 (2018).
Nguyen, A., Yosinski, J.,Clune, J. Deep neural networks are easily fooled: high confidence predictions for unrecognizable images, Preprint at https://arxiv.org/abs/1412.1897 (2014).
Geirhos, R., Janssen, D.H.J., Schütt, H.H., Rauber, J., Bethge, M., Wichmann, F.A. Comparing deep neural networks against humans: Object recognition when the signal gets weaker. Preprint at http://arxiv.org/abs/1706.06969 (2018).
Jones, T.D., Lawson, S.W., Benyon, D. & Armitage, A. Comparison of human and machine recognition of everyday human actions in Digital Human Modeling. ICDHM 2007. Lecture Notes in Computer Science, vol 4561 (ed. Duffy V.G.) 120–129 (Springer, 2007).
Fleuret, F. et al. Comparing machines and humans on a visual categorization test. Proc. Natl. Acad. Sci. U S A. 108(43), 17621–17625 (2011).
Dodge et al. A study and comparison of human and deep learning recognition performance under visual distortions. Preprint at https://arxiv.org/abs/1705.02498v1 (2017).
Finlayson, S. G. et al. Adversarial attacks on medical machine learning. Science 363(6433), 1287–1289 (2019).
Szegedy, C., et al. Intriguing properties of neural networks. Preprint at https://arxiv.org/abs/1312.6199v4 (2014).
Basu B.D. The Sacred Books of the Hindus vol VIII: The Nyaya Sutras of Gotama (ed. Basu, B.D.) 2 (1.1.4) (Panini Office, 1913); translation from Chadha, M. Perceptual Experience and Concepts in Classical Indian Philosophy. The Stanford Encyclopedia of Philosophy (Fall 2021 Edition). Edward N. Zalta (ed.), https://plato.stanford.edu/archives/fall2021/entries/perception-india (2021).
Fukushima, K. Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybernetics. 36, 193–202 (1980).
Lecun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 86(11), 2278–2324 (1998).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM. 60(6), 84–90 (2017).
Simonyan, K. Zisserman, A. Very deep convolutional networks for large-scale image recognition. Preprint at https://arxiv.org/abs/1409.1556v6 (2014).
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., Rabinovich, A. Going deeper with convolutions. Preprint at https://arxiv.org/abs/1409.4842v1 (2014).
Chollet, F., Xception: Deep learning with depthwise separable convolutions. Preprint at https://arxiv.org/abs/1610.02357 (2017).
Shaheed K. et al. DS-CNN: A pre-trained Xception model based on depth-wise separable convolutional neural network for finger vein recognition. Expert Syst. Appl. 191(C). https://doi.org/10.1016/j.eswa.2021.116288 (2022).
Yao, N. et al. L2MXception: An improved Xception network for classification of peach diseases. Plant Methods 17, 36 (2021).
He, K., Zhang, X., Ren, S. and Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778 (2016).
Tan, M., Le, Q.V. EfficientNet: Rethinking model scaling for convolutional neural networks. Preprint at https://arxiv.org/abs/1905.11946v5 (2019).
Touvron, H., Vedaldi, A., Douze, M., Jégou, H. Fixing the train-test resolution discrepancy: FixEfficientNet. Preprint at https://arxiv.org/abs/2003.08237 (2020).
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., Uszkoreit, J., Houlsby, N. An image is worth 16x16 Words: Transformers for image recognition at scale. Preprint at https://arxiv.org/abs/2010.11929 (2021).
Wortsman M., Ilharco G., Gadre S.Y., Roelofs R., Gontijo-Lopes R., Morcos A.S., Namkoong H., Farhadi A., Carmon Y., Kornblith S., Schmidt L. Model soups: Averaging weights of multiple fine-tuned models improves accuracy without increasing inference time. Preprint at https://arxiv.org/abs/2203.05482 (2022).
Chen T. & Guestrin C. XGBoost: A scalable tree boosting system. KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 785–794. https://doi.org/10.1145/2939672.2939785 (2016).
Kumar, M. & Kumar, M. XGBoost: 2D-Object Recognition Using Shape Descriptors and Extreme Gradient Boosting Classifier. In Computational Methods and Data Engineering Advances in Intelligent Systems and Computing, 1227 (eds Singh, V. et al.) (Springer, Singapore, 2021).
Chhabra, P., Garg, N. K. & Kumar, M. Content-based image retrieval system using ORB and SIFT features. Neural Comput. Appl. 32, 2725–2733 (2020).
Pizer, S. M. et al. Adaptive histogram equalization and its variations. Computer Vis. Graphics Image Process. 39, 355–368 (1987).
Garg, D., Garg, N. K. & Kumar, M. Underwater image enhancement using blending of CLAHE and percentile methodologies. Multimed. Tools Appl. 77, 26545–26561 (2018).
Shi J., Tomasi C. Good features to track. 9th IEEE Conference on Computer Vision and Pattern Recognition, 593–600 (1994).
Lowe, D. G. Object recognition from local scale-invariant features. Proc. Int. Conf. Computer Vis. 2, 1150–1157 (1999).
Bansal, M. et al. An efficient technique for object recognition using Shi-Tomasi corner detection algorithm. Soft Comput. 25, 4423–4432 (2021).
Gupta, S., Mohan, N. & Kumar, M. A study on source device attribution using still images. Arch. Comput. Methods Eng. 28, 2209–2223 (2021).
Patro, K. K. & Kumar, P. R. Machine learning classification approaches for biometric recognition system using ECG signals. J. Eng. Sci. Technol. Rev. 10(6), 1–8 (2017).
Patro, K. K., Jaya, P. A., Rao, M. J. & Kumar, P. R. An efficient optimized feature selection with machine learning approach for ECG biometric recognition. IETE J. Res. https://doi.org/10.1080/03772063.2020.1725663 (2020).
Phanikrishna, B. V., Jaya, P. A. & Suchismitha, C. Deep review of machine learning techniques on detection of drowsiness using EEG signal. IETE J. Res. https://doi.org/10.1080/03772063.2021.1913070 (2021).
Patro, K. K. et al. ECG data optimization for biometric human recognition using statistical distributed machine learning algorithm. J. Supercomput. 76, 858–875 (2020).
Rujano-Balza, M. A. Histology classifier app: Remote laboratory sessions using artificial neural networks. Med. Sci. Educ. 31(2), 1–3 (2021).
Mehra, R. Breast cancer histology images classification: Training from scratch or transfer learning?. ICT Exp. 4(4), 247–254 (2018).
Ahmed, S. et al. Transfer learning approach for classification of histopathology whole slide images. Sensors. 21, 5361 (2021).
The ImageMagick Development Team. ImageMagick. https://imagemagick.org/index.php (2021).
Eggert, P. shuf(1) Linux man page. https://www.unix.com/man-page/linux/1/shuf/ (2006).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Rosebrock, A. Transfer learning with Keras and deep learning. Pyimagesearch https://www.pyimagesearch.com/2019/05/20/transfer-learning-with-keras-and-deep-learning/ (2021).
Brownlee, J. Transfer learning in Keras with computer vision models. https://machinelearningmastery.com/how-to-use-transfer-learning-when-developing-convolutional-neural-network-models/ (2019).
Bradski, G. The OpenCV library. Dr. Dobb’s journal of software tools, https://opencv.org/ (2000).
Chollet, F., et al. Keras: The Python deep learning API. https://keras.io/ (2015).
Saxton, D., Grefenstette, E., Hill, F., Kohli, P. Analysing mathematical reasoning abilities of neural models. Preprint at https://arxiv.org/abs/1904.01557 (2019).
Liu, C. H., Collin, C. A., Burton, A. M. & Chaudhuri, A. Lighting direction affects recognition of untextured faces in photographic positive and negative. Vision. Res. 39(24), 4003–4009 (1999).
Elsayed, G.F., et al. Adversarial examples that fool both computer vision and time-limited humans. Preprint at https://arxiv.org/abs/1802.08195 (2018).
Russakovsky O., et al. ImageNet large scale visual recognition challenge. Preprint at https://arxiv.org/abs/1409.0575 (2015).
Funke, C.M., Borowski, J., Stosio, K., Brendel, W., Wallis, T.S.A, Bethge, M. Five points to check when comparing visual perception in humans and machines. Preprint at https://arxiv.org/abs/2004.09406v3 (2021).
Cowley, H. P. et al. A framework for rigorous evaluation of human performance in human and machine learning comparison studies. Sci. Rep. 12, 5444 (2022).
Rodriguez-Avi, J. et al. Methods for comparing two observed confusion matrices. Association of Geographic Information Laoratories in Europe, AGILE 2018 (conference poster). https://agile-online.org/conference_paper/cds/agile_2018/posters/96%20Poster%2096.pdf (2018).
Orosz, T. et al. Evaluating human versus machine learning performance in a LegalTech problem. Appl. Sci. 12, 297 (2022).
Kühl N., et al. Human vs. supervised machine learning: Who learns patterns faster? Preprint at https://arxiv.org/abs/2012.03661 (2020).
Acknowledgements
To the students and faculty of ACMS, Delhi Cantt, for participation and conduct of the study.
Author information
Authors and Affiliations
Contributions
S.B. & K.S.R. collected histological image data and reviewed their diagnosis. P.S. prepared the machine learning model and prepared image questionnaire for students. S.D. conducted the survey on medical students. A.M. planned the study, reviewed the histological images and designed the statistical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barui, S., Sanyal, P., Rajmohan, K.S. et al. Perception without preconception: comparison between the human and machine learner in recognition of tissues from histological sections. Sci Rep 12, 16420 (2022). https://doi.org/10.1038/s41598-022-20012-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20012-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.