Abstract
Computationally generated models of protein structures bridge the gap between the practically negligible price tag of sequencing and the high cost of experimental structure determination. By providing a low-cost (and often free) partial alternative to experimentally determined structures, these models help biologists design and interpret their experiments. Obviously, the more accurate the models the more useful they are. However, methods for protein structure prediction generate many structural models of various qualities, necessitating means for the estimation of their accuracy. In this work we present MESHI_consensus, a new method for the estimation of model accuracy. The method uses a tree-based regressor and a set of structural, target-based, and consensus-based features. The new method achieved high performance in the EMA (Estimation of Model Accuracy) track of the recent CASP14 community-wide experiment (https://predictioncenter.org/casp14/index.cgi). The tertiary structure prediction track of that experiment revealed an unprecedented leap in prediction performance by a single prediction group/method, namely AlphaFold2. This achievement would inevitably have a profound impact on the field of protein structure prediction, including the accuracy estimation sub-task. We conclude this manuscript with some speculations regarding the future role of accuracy estimation in a new era of accurate protein structure prediction.
Similar content being viewed by others
Introduction
Protein structure prediction (PSP) has been a major challenge in computational biology for half a century already1,2. Given a target protein sequence (hereafter referred to as ”target”), PSP methods aim to provide a three-dimensional model of the protein molecule. Such models help biologists build their theories and design their experiments. They provide a partial remedy to the high cost and much labor required for the experimental determination of structures. Typically, prediction methods generate many alternative structural models for each target. These models have diverse qualities even if generated by the same method, and often the best structural models of different targets are generated by different prediction methods. Unfortunately, large sets of alternative models do not provide much insight into biological problems, and the identification of the best models has been recognized early on3 as an essential PSP sub-task, known as Estimation of Model Accuracy (EMA, aka QA). EMA methods come in two flavors: local, assigning an accuracy measure to each residue of a model4,5,6,7, and global, assessing the qualities of complete models8,9. Often the former, in addition to its own merit, serves as a stepping stone to the latter10,11,12.
This manuscript presents MESHI_consensus, a new EMA method, with state-of-the-art performance. Specifically, MESHI_consensus aims to predict the similarity of protein models to the corresponding native structures in terms of the zero to one Global Distance Test Total Score (GDT_TS), which assigns a score of one to models that are very similar to the native structure and lower scores to models that are less similar. A score close to zero indicates an irrelevant model.
In the last two decades, the Critical Assessment of Protein Structure Prediction (CASP), a biannual and community-wide series of prediction experiments13,14,15 monitor the performance of prediction methods and accelerate their development. In each experiment, CASP organizers collect around a hundred targets at the final stages of their structural determination, and challenge researchers to submit blind predictions of the yet unknown structures. The assessment of these models, when the structures are finally determined, allows a reliable evaluation of prediction methods. Over the years CASP has become the de facto ”Gold Standard” of the PSP field, and the recent unprecedented performance of AlphaFold2 in 14th CASP experiment16,17 is commonly recognized as marking a new era in structural biology. The CASP experiments have several tracks for PSP sub-tasks, and since 2008, EMA is considered a CASP category8,18,19,20,21. The relevance of EMA in an era of high accuracy structural modeling is considered in the Discussion section below. CASP experiments play a two-fold role in the current study: our benchmark22 is based on structural models submitted to the 9th to 13th CASP rounds, and the 14th experiment is used to evaluate the new EMA method.
Since the early days of EMA23,24 and up until recently, the best performing methods have used the consensus (aka multi-model) approach, which considers structural similarity between independently generated models as an indication that they are likely to be all similar to the unknown native structure8,18,19,25. Not withstanding their power however, consensus EMA does not provide any insight about the actual physics of protein folding or the essence of being a correct structure. Further, it cannot be applied to a single model, and fails to identify exceptionally good models. These limitations motivate an alternative, single-model, approach that considers the internal properties of a single model structure (e.g., estimated energy and compactness) as well as its compatibility with one dimensional predictions of secondary structure and solvent accessibility, which are based of multiple sequence alignments (MSAs)26,27,28,29. Recently, compatibility with contact predictions derived from deep multiple sequence alignments seems to be a game changer, considerably improving EMA performance and allowing single-model methods to outperform consensus based ones21,30,31.
Most recent EMA methods use machine learning (ML) algorithms, including neural networks32,33,34, SVM35,36,37, and tree-based methods38, to create a statistical model that combines measurable features into a single number, which estimates the quality of a structural model35,39. To this end, ML algorithms use datasets of annotated structural models and learn the intricate relations between the features and model quality. Specifically, the EMA methods use model structures to produce meaningful features, such as statistical pairwise potentials40,41 and consensus-derived terms42. These features constitute the input for regression models9,43 that integrate them into a single score. An emerging Deep learning-based approach eliminates the distinction between feature generation and and learning of statistical model. It uses convolutional6,44,45 and graph46,47 neural networks to derive the scores directly from elementary features such as atom/residue types and distances. Higher order features, analogous to energy terms and other traditional features, emerge as information flows throw the the networks layers.
Our method, MESHI_consensus, uses a tree-based machine-learning algorithm to estimate model qualities from 982 structural and consensus features.
Methods
The following sections introduce the basic components of MESHI_consensus. We first present our benchmark, a dataset of targets and structural models thereof from previous CASP experiments, and the features derived from them. Then we describe the performance measures that guided the development of the ML model, as well as the model design procedure, which includes regressor and hyper-parameters selection. Finally, we present a data filtering process that reduces training set noise.
Structural models dataset
We trained and evaluated our method using a dataset that consists of 73,053 single-chain models, which were generated as blind server predictions of 345 CASP9-CASP13 targets (2010–2018) (Table S1) an extension of the dataset used in22. These targets are a non-redundant subset of the \(\approx\)430 targets of these CASP experiments. To this end, targets were considered redundant, and discarded, if a newer target was strictly similar by either sequence (\(E-value < 10^{-3}\)) or structure (more than a half of the residues could be structurally aligned by the iterative magic fit method of Swiss-PDB-Viewer48,49). Duplicated models, having identical conformations as other ones (typically from different servers of the same group), were identified and removed.
Many server-generated models include clashes (too short distances) between atoms and other structural distortions, such as deviations from correct bond lengths and angles. Thus, before feature extraction, each structural model was subjected to energy minimization using the MESHI molecular modeling package50. The energy function includes strong spatial constraints, and most distortions are removed with negligible structural changes.
The test set of this study includes 67 CASP14 targets (10,889 structural models), which were predicted by modeling servers during the CASP14 experiment (May–August, 2020). Both model generation and the estimation of their accuracy by MESHI_consensus web server were done in a blind fashion before their structures became available. After the models were downloaded from the CASP14 website they were energy minimized by MESHI package and their features were fed to the MESHI_consensus model.
Features dataset
In this study, each structural model is represented by a vector of 982 features. These features may be divided into two broad classes: structural model-features, and target-features that modulate the former.
Structural model-features
The following features are calculated using the MESHI package50.
-
Basic features: 142 features derived solely from single model structures51. They include a mix of commonly used and novel knowledge-based energy terms (some of which are unpublished yet). These terms include pair-wise atomic potentials40,52,53, torsion angles54, hydrogen bonds, and hydrogen bond patterns55, solvation terms, ”meta” energy terms that consider the distribution of other terms within the protein atoms, an extended radius of gyration that takes into account different classes of amino acids (polar vs. non-polar, secondary structure elements vs. coil region, etc.), and compatibility of the models with solvent exposure prediction56, and with 3-, 8- and 13-classes secondary structure predictions57,58,59.
-
Consensus features: seven features (Eqs. 1–7) that represent the similarities between a model of a specific target and the other models of the same target. These terms are calculated as follows: \(\forall d \varvec{\in } T\), where T is the set of structural models of some target.
$$\begin{aligned} g d t_{i}\_\text{consensus }(d)=\frac{1}{|T|} \sum _{s \in T} g d t_{i}(d, s) \\ \text{ where } \text{ i } \in \{{\mathbf {1}}, {\mathbf {2}}, {\mathbf {4}}, {\mathbf {8}}\} \end{aligned}$$(1)$$\begin{aligned} g d t_{-} t s_{-} \text{consensus }(d)=\frac{1}{|T|} \sum _{s \in T} g d t_{-} t s(d, s) \end{aligned}$$(2)$$\begin{aligned} g d t_{-} h a_{-} \text{consensus }(d)=\frac{1}{|T|} \sum _{s \in T} g d t_{-} h a(d, s) \end{aligned}$$(3)$$\begin{aligned} r m s_{-} \text{consensus }(d)=\frac{1}{|T|} \sum _{s \in T} R M S D(d, s) \end{aligned}$$(4)Where: \(\forall d, s \varvec{\in } T\)
$$\begin{aligned} g d t_{j \in \{0.5,1,2,4,8]}\left( d, s\right) = \text{ The}\, \text{maximal} \,\text{fraction}\, \text{of} \, {\varvec{s}}\, \text{residues}\, \text {that} \, \text{are} \, \text {less} \, \text{than} \,j\, {{^\circ\!\!\,\,\!\!\!{\rm A}} }\, \text {form} \,\text {the} \, \text {corresponding} \, \text {residues} \, \text {of} \, {\varvec{d}} \,\text{after} \, \text {superposition}. \end{aligned}$$(5)$$\begin{aligned} g d t_{-} t s(d, s)=\frac{\sum _{i \in \{1,2,4,8\}} g d t_{i}(d, s)}{4} \end{aligned}$$(6)$$\begin{aligned} g d t_{-} h a(d, s)=\frac{\sum _{k \in \{0.5,1,2,4\}} g d t_{k}(d, s)}{4}\\ \end{aligned}$$(7)
Target-features
EMA datasets are organized in two levels; the objects that we study, and whose accuracies we predict are structural models. Yet each model belongs to a specific target (with no overlap between the targets). Each target is characterized by a unique sequence, which is shared by all its models, and a unique native structure. Thus, the mapping of features to model qualities may be biased by target characteristics such as length and chemical composition (amino acid sequence), which differ between targets but are typically identical in most models of a given target. Therefore, feature distributions and their relation to model quality differ between targets (Fig. 1). Further, some training set targets may be less informative than others, with respect to certain features, due to specific characteristics such as ligand binding.
Target characteristics bias feature values. Each point in the figure represents the ”GOAP”S energy60 and accuracy of a single structural model. Models of the two CASP targets T0601 and T0530 are depicted by blue and red points respectively. Accuracy is measured in GDT_TS between the models and the native structures, that is \(g d t_{-} t s(d, n)\), Eq. (6), where n is the native structure of that target). ”GOAP” energy term is a strong feature and it (anti) correlates well with the accuracies of both targets. Yet, given a feature value range (e.g., around–10,000), the qualities of the two proteins are very different.
We use this domain knowledge to generate target-specific features that allow the learning process to modulate the outcome of the model features:
-
One-hot encoding of target name: binary features (one per target). That is, for each target T of the training set, there is a feature \(OH_T\) such that \(OH_T = 1\) for all the models of T and 0 otherwise. When positioned in a node of a decision tree, \(OH_T\) splits the leaves of the sub-tree to T and \(non-T\), rendering the features in the nodes of the T sub-tree practically meaningless. Interestingly the training process does make use of this ability to eliminate the effect of specific features in specific targets.
-
Z-score, a normalized (zero mean and standard deviation of one) version of each basic feature, based on the target’s mean and standard deviation.
-
Amino acid composition:
-
20 features for the frequency of each amino acid in the sequence of the target.
-
6 features for the frequency of amino acids with certain properties in the sequence: Positive charged, negative charged, aromatic, polar, and non-polar.
-
Combining all the feature vectors of the structural models dataset creates a features dataset.
Performance measures
We aim to predict the accuracy of structural models in terms of GDT_TS between the models and the native structures, that is \(g d t_{-} t s(d, n)\) (Eq. 6) where n is the native structure of that target. Specifically, we use three performance criteria:
-
1.
Root mean square of prediction errors (RMSE)—the per-target distance between the prediction values and the observed (true) values.
-
2.
LOSS - for each target, the difference between the quality of the best model (highest observed GDT_TS) and the quality of the top-ranking model.
-
3.
5-LOSS - for each target, the minimum difference between the quality of the best model (highest observed GDT_TS) and the qualities of the five top-ranking models.
For dataset models, method performance is estimated by the median of 345 Leave-One-Target-Out cross-validation experiments (one per dataset target). In each experiment, the statistical model is trained using all the targets except one, which serves as the test set. This strategy is computationally expensive but reduces biases, and in a sense simulates the real-world scenario, where we learn from all the models of targets whose native structures are known and assess the model qualities of a target whose structure is yet unknown.
Method design
The design of these EMA methods aimed to optimize two performance criteria: the median of the per-target RMSE and median LOSS. We used Leave-One-Target-Out cross-validation experiments to choose the regressor, its hyper-parameters, and the data filtering strategy.
Regressor
In this work, we formulate EMA as a regression problem that maps measurable features of the structural models to a continuous quality score (GDT_TS) between the models and the native structures. To this end, we tested three regressors: linear regression, Light Gradient Boosted Machine (LightGBM) regressor61, and a fully connected neural network. The superior performance (Fig. 2) of LightGBM motivated us to examine five other tree-based regressors: BaggingRegressor, GradientBoostingRegressor, RandomForestRegressor, and ExtraTreesRegressor from the scikit-learn library62, as well as Extreme Gradient Boosting (XGB) regressor63. We remained with LightGBM, however, as it outperforms all five by a small margin and is faster to train.
LightBGM61 outperforms linear regression and neural networks. The box plots depict the results of Leave-One-Target-Out experiments in terms of RMSE (A) and LOSS (B), using three different regressors. The difference in performance between LightBGM and the two other methods is statistically significant (Wilcoxon one-sided test with a p-value < 1.39e−51). The performances of few other tree-based methods were practically indistinguishable from LightBGM (data not shown) but computation time was much longer.
Hyperparameters
We used grid search to find the optimal values for two hyperparameters of LightGBM regressor: learning rate and the number of estimators. Learning rate 0.1 and 100 estimators achieved good results in a reasonable computation time. For all the other parameters, we used the default values as supplied by the LightGBM framework. Specifically, the loss function that the regressor training minimizes is the RMSE.
Data filtering
Some of the dataset targets are isolated chains of multi-subunit complexes (e.g., single helices of helix-bundles). Estimating their quality is a challenge due to hydrophobic interface residues that are superficially exposed when seen out of context. For such targets, MESHI may produce feature values that are inconsistent with the label (GDT_TS), increasing noise and impairing the learning process. The qualities of such targets are hard to estimate, even in an over-fitting scenario. The removal of 18 such targets (Table S1) from the training set significantly reduces the median error of quality estimates and does not affect the identification of the best models (Fig. 3).
Filtering out outlier targets from the training set reduces the error in quality estimation (A), and does not affect the identification of the best models (B). The box plots depict the results of Leave-One-Target-Out experiments with and without data filtering in the training set. (A) The median of the RMSE is significantly reduced by data filtering (Wilcoxon one-sided test, with a p-value of 0.005). (B) Data filtering does not affect the distribution of LOSS (the quality differences between the top-ranking model, and the best model in the set). Many of the worse performing outliers in the plots are proteins that were filtered out from the training set.
Results
MESHI_consensus was developed using a dataset of CASP server models that were generated as blind predictions in five consecutive CASP experiments (9–13). CASP14 models serve as the ultimate test set as their true qualities were unknown at the time of prediction. Here we present the method’s performance in predicting the accuracies of models in the dataset, consider the contributions of different feature types, and conclude by presenting, and discussing CASP14 performance.
Dataset performance
The performance of MESHI_consensus is estimated by Leave-One-Target-Out experiments on the structural models dataset. The qualities of about half of the targets are estimated well (Fig. 4), with small (< 0.05) RMSE, and low (<0.04) LOSS. A small fraction of the targets (< 6%) are practically missed, with RMSE or LOSS above 0.2.
Benchmark performance of MESHI_consensus. The results of 345 leave-one-target-out experiments are summarized in three histograms: (A) RMSE; (B) LOSS, the difference between the quality of the top-ranking model and the quality of the best one. (C) LOSS5, the minimal difference between the qualities of 5 top-ranking models and the quality of the best one. Median (red) and mean (green) values are indicated by vertical lines.
Feature importance
MESHI_consensus uses a large number of features for the estimation of model accuracies. To assess their contributions to the LBGM statistical model, we first checked the importance ranking of the basic features and all the features. Following the results (Fig. 5), we also performed Leave-One-Target-Out experiments with only the basic features, only the consensus features, and with all the features (Fig. 6). Additional subgroups of features were tested (data not shown), but the best result was obtained when we used the whole set of features. The measure we used for the importance of the features is ”GAIN”, which is the sum of the information gains of all splitting points that use that feature, where information gain is the Kullback–Leibler64 divergence of the data before and after the split. A higher value of this metric, when compared to another feature, implies it is more important for the predictive model.
The ten highest importance features in LightGBM models that use either all the features or only the basic ones are depicted in Fig. 5. These models were trained on all the benchmark targets. Qualitatively similar estimates of feature GAINs, derived from benchmark subsets are reported in Feature importance file (supplementary material). All but one of the LightGBM models make use of the possibility to distinguish between structural models from different targets. When target-specific features are available to the models, they choose from a wide variety, without a clear preference. The basic features were not intended to include such features explicitly, yet the models assign relatively high importance to the total number of side-chain atoms in the model. This feature seems almost arbitrary and was added to the basic features by mistake (being a component of other features). We speculate that the models consistently chose it as it allows target distinction. When all features are considered, consensus features are ranked highest by a large margin, apparently, because good (i.e, close to native) models are similar to one another (they are all similar to the native structure), while low-quality models can be very different from each other. This observation is consistent with the dominance of consensus-based methods in the EMA field. When only the basic features are considered, two classes of features become dominant. The first class (e.g., sasaCompatibility), quantifies the agreement between the observed and predicted solvent exposed area and the secondary structure of model residues. Apparently, an inability to reproduce them is a strong indicator of low model quality. This is consistent with the disruptive effect of out-of-context targets (e.g., isolated subunits) on learning.
The second important class of basic features (e.g., goap_ag) rewards structural traits that are common in native structures. One common trait is compactness which manifests itself by a large number of atom contacts. The other common trait is the abundance of specific atomic interactions (e.g., contacting side-chain atoms of hydrophobic residues). Having many favorable interactions, and a compact structure are strong indicators of high model quality. Finally, as our quality measure, GDT_TS between the models and the native structures represents the fraction of accurately modeled residues, it is bound by the fraction of the target sequence which is actually modeled. Coverage features depict this fraction and are consistently ranked among the ten most important.
Considering the dominance of consensus features, we tested whether the other features are needed at all. To this end, we performed a Leave-One-Target-Out experiment with three different sets of features (Fig. 6). The first experiment served as a baseline and included only the basic features. The second experiment used only the consensus features and the third used all the features. In both, the RMSE and LOSS, the best results are obtained by using the entire set of features. For RMSE (A), which is the loss function of the regressor, most of the improvement is due to the consensus features, consistent with the ”GAIN” results (Fig. 5). Yet by using all the features we obtain statistically better performance. For LOSS (B), the basic features outperform the consensus ones, reflecting the difficulty of consensus features to identify exceptionally good models. The best structural model of target T0581 for example (BAKER-ROSETTASERVER_TS4, 0.64 GDT_TS) was picked by the statistical model that was trained with the basic features only. Training with all the features resulted picking the second-best model (GDT_TS 0.33), which is similar to some other inaccurate ones. Notably, however, adding consensus features to the basic ones reduces the number of outliers and the magnitude of their deviation from the median performance. This is probably because consensus features are indifferent to complex subunits and membrane proteins that distort many basic features.
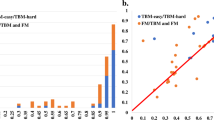
The ten most important (highest “GAIN”) features considering only the basic features (A) and all features (B). The features from top to bottom: sasaCompatibility—a measure of the agreement between the solvent accessible surface area of the model’s residues (as measured by DSSP65) and their predicted accessibility56. goap_ag—a pairwise orientation-dependent knowledge-based potential40. deepCNF8Compatibility—a measure of the agreement between the secondary structure (8 states) of the model’s residues (as measured by DSSP65) and their predicted secondary structure66. contacts8 and contacts14—the average numbers of contacts with thresholds of 8 Å and 14 Å, respectively, between carbon atoms. meshinr_dssp8Compatibility and meshirw_dssp8Compatibility_Weighted—two slightly different measures of the agreement between the secondary structure (8 states) of the model’s residues (as measured by DSSP65) and their predicted secondary structure59. scCarbonN—the number of carbon atoms in the model’s side-chains. coverage—the fraction of the target sequence, which the are modeled. SheetFraction—the fraction of beta-sheet resides within the residues with any secondary structure. consensus features—see Eqs. (1–4). gdt1_consensus_median—the median value of gdt1_consensus, among all the models of a specific target. hydrogenBondsPairs_median—the median value of a cooperative hydrogen bonds energy term67 among all the models of a specific target. cooperativeZstdRamachandranSidechain_median—the median value of a cooperative torsion angle energy term, among all the models of a specific target.
Per-target performance with different feature sets. The box plots show the per-target performance in terms of RMSE (A) and LOSS (B), of Leave-One-Target-Out experiments with the basic features (blue), consensus features (orange), and all features (green). The differences between the median performances of all the features and the median performances of the feature subsets are statistically significant (Wilcoxon one-sided test with a p-value <10e−4).
CASP14: MESHI_consensus
The previous round of CASP experiments, CASP14 (May–August, 2020) serves as the independent test set of this study. MESHI_consensus took part in that experiment as an EMA server and submitted 11,135 global quality predictions of server models.
One target, T1093, was missed due to technical failure. Figure 7 depicts the best (left) and worst (right) results with targets T1046s2 and T1031 respectively. The overall performance on this test set (Fig. 8) is comparable to that of the benchmark (Fig. 4). The slight performance reduction is discussed below. Comparison with the other 71 research groups that competed in the EMA category reveals a state-of-the-art performance, with more than 65% of the predictions ranked within the top 10 in either LOSS or RMSE. Specifically, MESHI_consensus reached:
-
Top GDT_TS MCC(50) score (Table 1A)
-
Third lowest average prediction error (Table 1B)
-
Sixths lowest average LOSS (Table 1C)
Notably, among the other top-performing methods, one (MESHI) is a curiosity-driven variant of MESHI_consensus variant, that simply adds server names as a feature and ranked a bit higher.
Notwithstanding these achievements, the overall performance of MESHI_consensus in CASP14 (Fig. 8) is worse than in the dataset’s Leave-One-Target-Out experiments (Fig. 4). Notably, nine of the EMA targets are domains of a single large protein (Fig. 9). MESHI_consensus failed to predict six of them with LOSS values ranging from 0.16 to 0.4 (Fig. 7, right). We speculate that the lack of the protein context had to do with the poor performance, as no other targets showed so high LOSS values. As demonstrated in Fig. 9, these structures have numerous inter-domain stabilizing interactions, that are missing in the isolated EMA targets. A similar phenomenon is also observed in the dataset (see the Data filtering section). Target T1073 also showed exceptionally bad performance with an RMSE of 0.41. Unfortunately, we cannot study this case, as its structure has not been published yet. Another, more speculative explanation for the lower performance is the methodological turning point of CASP14 (discussed below). It raises a major challenge to MESHI_consensus, as well as to any supervised learning method that uses sets of CASP server models training. The test (CASP14 models) and training sets were not sampled from the same distribution, as the models of CASP14 are on average more accurate than those of previous CASP experiments.
Examples of MESHI_consensus success and failure in CASP14. Predicted vs. observed qualities of models from two targets: Left: For target T1046s2 (PDB code: 6px4), MESHI_consensus reached a low RMSE (0.074) and the top-scoring model is indeed the best one (zero LOSS). Right: For target T1031 (PDB code: 6vr4) the RMSE is 0.128 and the best model ranked very low (LOSS is 0.428)
Three examples of apparent bias of EMA prediction, by the absence of molecular context. The figure presents the large (2225 residues) DNA-dependent RNA polymerase of crAss-like phage phi14:2 (PDB 6VR4) that gave rise to ten CASP14 targets: The whole protein (T1044), which was not offered as an EMA target, and nine domain targets T1031, T1033, T1035, T1037, T1039, T1040, T1041, T1042, and T1043. For six of them: T1031, T1033, T1035, T1039, T1040, and T1043, MESHI_consensus failed to provide reliable EMA predictions (LOSS above 0.16). We speculate that these failures may be attributed, at least partially, to the absence of the protein context in the isolated domains. (a) The three domain targets, depicted in the context of the whole protein (yellow): T1031 (residues 1–95, red), T1033 (residues 96–196, cyan), T1040 (residues 1372–1501, magenta). (b–d) The three interfaces of these domains (respectively) with the rest of the protein. A sample of the interacting residues is shown, the rest are hidden for clarity. Blue dashed lines represent hydrogen bonds and salt bridges (Figure is drawn with ChimeraX68).
Model accuracy with respect to two alternative native structures. Target T1025 was evaluated by the CASP14 assessors with the ligand-bound D-glucose O-methyltransferase as its native structure (PDB entry 6UV669). The structure of the unbound protein is also available (PDB entry 6UVQ70). While the choice of either structure is arbitrary, the resulted performance measures are quantitatively different. A theoretical EMA oracle that provides 6UVQ based accuracy as its prediction, would have 4% RMSE and 6% LOSS. The dots represent server models, the black line is the diagonal (x \(=\) y), and the red and magenta circles depict the top model by 6UV6 and 6UVQ respectively.
Discussion
This manuscript introduces MESHI_consensus, a new method for quality assessment of protein models. MESHI_consensus uses a large and diverse set of features, representing both physical concepts (e.g., energy terms) and domain knowledge (target-specific and consensus-based features). The features are integrated to a single score by a powerful and computationally efficient, tree-based LightGBM regressor61. One type of state-of-the-art features, which the method lacks, is compatibility with predicted distances derived from multiple sequence alignments (MSA)11,76. MSA-driven distances are the keystone of the current breakthrough in PSP, and compatibility with them seems to be a powerful feature20.
The development of MESHI_consensus was guided by Leave-One-Target-Out experiments using a non-redundant dataset of structural models from previous CASP experiments. The recent CASP14 provided an objective performance test and MESHI_consensus scores among the top methods (see Results). One may speculate that had we used contact compatibility features we could perform better. During the study, we invested much effort in analyzing failures, that is targets for which we considerably missed the actual model qualities and/or their rankings. Many of these failures could be rationalized in retrospect as related to the inability of our features to consider stabilizing inter-molecular interactions. We tried to implement insights from this analysis through data filtering with limited success. One may hope though that a more systematic approach to this problem may lead to better performance in future studies.
A profound limitation of MESHI_consensus, is the reliance on a single native structure as the gold standard for the labeling of the data. This is in line with the common practice in the field, which is applied in CASP and in all the studies that we are aware of. Yet, this practice ignores the structural flexibility of proteins as manifested by diverse structures of the same protein in different PDB entries77, and in the results of NMR studies. Figure 10 demonstrates this observation by assessing the server models of target T1025 using the ligand-bound and apo crystal structures of that protein. Target T1064 shows a similar, yet less pronounced trend (data not shown). In the CASP context alternative structures are rarely available, and thus ignoring them is practically unavoidable, and can be seen as part of the ”noise” characteristic of any experiment. In the training phase, however, ignoring available knowledge of structural multiplicity adds superficial, arbitrary, constraints to the learning process, and probably harms the resulted statistical model. Structural multiplicity can be introduced into the training phase of EMA methods if structural models are evaluated by their similarity to the closest of the known structures, rather than to a single arbitrary one. We have already demonstrated the usefulness of a similar approach in the related fields of secondary structure prediction78 and knowledge-based energy functions79.
Finally, CASP14 has witnessed a remarkable breakthrough in PSP, with many models of hard targets reaching experimental quality. This achievement had a limited effect on the EMA section of CASP14 as the cutting-edge method, AlphaFold16, did not provide a server. Yet, it is evident that a new standard of model qualities is set80. Will EMA be needed at all when the models are ”almost perfect”? An obvious answer is that we are in the middle of the event and its consequences cannot be predicted. More fundamentally, the new achievements seem to open new horizons for PSP, considerably improving our ability to cope with essential problems like structures of molecular complexes and protein dynamics. These challenges require their own EMA tools.
We believe that the insights of this study, most importantly the central role of structural multiplicity and molecular context, will gain much importance in the era of high-accuracy modeling. On a more speculative note, we suggest that features like the ones used in this and related studies will also remain relevant, as design principles for new, probably neural-network-based, architectures. The current application of neural network techniques to EMA use standard architectures and avoid ”feature engineering”, such as energy terms. One may speculate that introducing more domain knowledge into the network architecture and its input features will result in more accurate and stable performance.
Data availability
The data sets generated and analysed during the current study are available at http://meshi1.cs.bgu.ac.il/BittonAndKeasar2021/.
References
Levitt, M. & Warshel, A. Computer simulation of protein folding. Nature 253, 694–698. https://doi.org/10.1038/253694a0 (1975).
Zwanzig, R., Szabo, A. & Bagchi, B. Levinthal’s paradox. Proc. Natl. Acad. Sci. 89, 20–22 (1992).
Ginalski, K., Elofsson, A., Fischer, D. & Rychlewski, L. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018. https://doi.org/10.1093/bioinformatics/btg124. https://academic.oup.com/bioinformatics/article-pdf/19/8/1015/642841/btg124.pdf (2003).
Wallner, B. & Elofsson, A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 15, 900–913 (2006).
Studer, G., Biasini, M. & Schwede, T. Assessing the local structural quality of transmembrane protein models using statistical potentials (qmeanbrane). Bioinformatics 30, i505–i511 (2014).
Takei, Y. & Ishida, T. P3cmqa: Single-model quality assessment using 3dcnn with profile-based features. Bioengineering 8, 40 (2021).
Shuvo, M. H., Bhattacharya, S. & Bhattacharya, D. Qdeep: Distance-based protein model quality estimation by residue-level ensemble error classifications using stacked deep residual neural networks. Bioinformatics 36, i285–i291 (2020).
Wallner, B. & Elofsson, A. Prediction of global and local model quality in CASP7 using Pcons and ProQ. Proteins Struct. Funct. Bioinform. 69, 184–193. https://doi.org/10.1002/prot.21774 (2007) (number: S8).
Mirzaei, S., Sidi, T., Keasar, C. & Crivelli, S. Purely structural protein scoring functions using support vector machine and ensemble learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 1515–1523. https://doi.org/10.1109/TCBB.2016.2602269 (2019).
Uziela, K. & Wallner, B. Proq2: Estimation of model accuracy implemented in Rosetta. Bioinformatics 32, 1411–1413 (2016).
Maghrabi, A. H. & McGuffin, L. J. Modfold6: An accurate web server for the global and local quality estimation of 3D protein models. Nucleic Acids Res. 45, W416–W421 (2017).
Olechnovic, K. & Venclovas, C. Voromqa: Assessment of protein structure quality using interatomic contact areas. Proteins Struct. Funct. Bioinform. 85, 1131–1145. https://doi.org/10.1002/prot.25278. https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.25278 (2017).
Moult, J. A decade of CASP: Progress, bottlenecks and prognosis in protein structure prediction. Curr. Opin. Struct. Biol. 15, 285–289. https://doi.org/10.1016/j.sbi.2005.05.011 (2005).
Moult, J., Fidelis, K., Kryshtafovych, A., Rost, B. & Tramontano, A. Critical assessment of methods of protein structure prediction—Round VIII. Proteins Struct. Funct. Bioinform. 77, 1–4. https://doi.org/10.1002/prot.22589 (2009) (number: S9).
Moult, J., Fidelis, K., Kryshtafovych, A., Schwede, T. & Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)—Round x. Proteins Struct. Funct. Bioinform. 82, 1–6. https://doi.org/10.1002/prot.24452. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.24452 (2014).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 1–11. https://doi.org/10.1038/s41586-021-03819-2 (2021).
Pereira, J. et al. High-accuracy protein structure prediction in CASP14. Proteins Struct. Funct. Bioinform. (John Wiley & Sons, Ltd.) 89, 1687–1699 (2021).
Kryshtafovych, A. et al. Assessment of the assessment: Evaluation of the model quality estimates in CASP10. Proteins Struct. Funct. Bioinform. 82, 112–126. https://doi.org/10.1002/prot.24347. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.24347 (2014).
Kryshtafovych, A., Monastyrskyy, B., Fidelis, K., Schwede, T. & Tramontano, A. Assessment of model accuracy estimations in CASP12. Proteins Struct. Funct. Bioinform. 86, 345–360. https://doi.org/10.1002/prot.25371 (2018). (number: S1).
Cheng, J. et al. Estimation of model accuracy in casp13. Proteins Struct. Funct. Bioinform. 87, 1361–1377 (2019).
Kwon, S., Won, J., Kryshtafovych, A. & Seok, C. Assessment of protein model structure accuracy estimation in CASP14: Old and new challenges. Proteins Struct. Funct. Bioinform. (John Wiley & Sons, Ltd.) 89, 1940–1948 (2021).
Sidi, T. & Keasar, C. Loss-functions matter, on optimizing score functions for the estimation of protein models accuracy. bioRxiv 651349 (2019).
Lundström, J., Rychlewski, L., Bujnicki, J. & Elofsson, A. Pcons: A neural-network-based consensus predictor that improves fold recognition. Protein Sci. 10, 2354–2362 (2001).
Ginalski, K., Elofsson, A., Fischer, D. & Rychlewski, L. 3d-jury: A simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018 (2003).
Kryshtafovych, A., Fidelis, K. & Tramontano, A. Evaluation of model quality predictions in CASP9. Proteins Struct. Funct. Bioinform. 79, 91–106. https://doi.org/10.1002/prot.23180 (2011). (number: S10).
Wallner, B. & Elofsson, A. Can correct protein models be identified?. Protein Sci. 12, 1073–1086 (2003).
Mirzaei, S., Sidi, T., Keasar, C. & Crivelli, S. Purely structural protein scoring functions using support vector machine and ensemble learning. in IEEE/ACM Transactions on Computational Biology and Bioinformatics. 1–1. https://doi.org/10.1109/TCBB.2016.2602269 (2016). (number: 99).
Olechnovič, K. & Venclovas, Č. Voromqa: Assessment of protein structure quality using interatomic contact areas. Proteins Struct. Funct. Bioinform. 85, 1131–1145 (2017).
Ray, A., Lindahl, E. & Wallner, B. Improved model quality assessment using proq2. BMC Bioinform. 13, 1–12 (2012).
McGuffin, L. J., Aldowsari, F. M. F., Alharbi, S. M. A. & Adiyaman, R. ModFOLD8: Accurate global and local quality estimates for 3D protein models. Nucleic Acids Res. 49, W425–W430. https://doi.org/10.1093/nar/gkab321 (2021).
Hiranuma, N. et al. Improved protein structure refinement guided by deep learning based accuracy estimation. Nat. Commun. 12, 1–11 (2021).
Faraggi, E. & Kloczkowski, A. A global machine learning based scoring function for protein structure prediction. Proteins Struct. Funct. Bioinform. 82, 752–759. https://doi.org/10.1002/prot.24454. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.24454 (2014).
Zhang, J. & Xu, D. Fast algorithm for population-based protein structural model analysis. Proteomics 13, 221–229. https://doi.org/10.1002/pmic.201200334 (2013). (number: 2).
Terashi, G., Nakamura, Y., Shimoyama, H. & Takeda-Shitaka, M. Quality assessment methods for 3D protein structure models based on a residue–residue distance matrix prediction. Chem. Pharmaceut. Bull. 62, 744–753 (2014).
Qiu, J., Sheffler, W., Baker, D. & Noble, W. S. Ranking predicted protein structures with support vector regression. Proteins Struct. Funct. Bioinform. 71, 1175–1182. https://doi.org/10.1002/prot.21809 (2008). (number: 3).
Manavalan, B. & Lee, J. SVMQA: Support-vector-machine-based protein single-model quality assessment. Bioinformatics 33, 2496–2503. https://doi.org/10.1093/bioinformatics/btx222 (2017). (number: 16).
Hippe, K., Lilley, C., Berkenpas, W., Kishaba, K. & Cao, R. Zoomqa: Residue-level single-model QA support vector machine utilizing sequential and 3D structural features. bioRxiv (2021).
Manavalan, B., Lee, J. & Lee, J. Random forest-based protein model quality assessment (RFMQA) using structural features and potential energy terms. PloS one 9, e106542 (2014).
Wang, Z., Tegge, A. N. & Cheng, J. Evaluating the absolute quality of a single protein model using structural features and support vector machines. Proteins Struct. Funct. Bioinform. 75, 638–647. https://doi.org/10.1002/prot.22275 (2009). (number: 3).
Zhou, H. & Skolnick, J. GOAP: a generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys. J. 101, 2043–2052 (2011). (number: 8).
Zhang, J. & Zhang, Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PloS one 5, e15386 (2010).
Lundström, J., Rychlewski, L., Bujnicki, J. & Elofsson, A. Pcons: A neural-network-based consensus predictor that improves fold recognition. Protein Sci. 10, 2354–2362. https://doi.org/10.1110/ps.08501 (2001) (number: 11).
Korovnik, M. et al. Synthqa-hierarchical machine learning-based protein quality assessment. bioRxiv (2021).
Derevyanko, G., Grudinin, S., Bengio, Y. & Lamoureux, G. Deep convolutional networks for quality assessment of protein folds. Bioinformatics 34, 4046–4053, https://doi.org/10.1093/bioinformatics/bty494. https://academic.oup.com/bioinformatics/article-pdf/34/23/4046/26676600/bty494.pdf (2018).
Pagès, G., Charmettant, B. & Grudinin, S. Protein model quality assessment using 3D oriented convolutional neural networks. Bioinformatics 35, 3313–3319. https://doi.org/10.1093/bioinformatics/btz122 (2019).
Sanyal, S., Anishchenko, I., Dagar, A., Baker, D. & Talukdar, P. Proteingcn: Protein model quality assessment using graph convolutional networks. bioRxivhttps://doi.org/10.1101/2020.04.06.028266. https://www.biorxiv.org/content/early/2020/04/07/2020.04.06.028266.full.pdf (2020).
Baldassarre, F., Menéndez Hurtado, D., Elofsson, A. & Azizpour, H. GraphQA: Protein model quality assessment using graph convolutional networks. Bioinformatics 37, 360–366. https://doi.org/10.1093/bioinformatics/btaa714 (2021).
Kaplan, W. & Littlejohn, T. G. Swiss-pdb viewer (deep view). Brief. Bioinform. 2, 195–197 (2001).
Guex, N. & Peitsch, M. C. Swiss-model and the swiss-pdb viewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Kalisman, N. et al. MESHI: A new library of Java classes for molecular modeling. Bioinformatics 21, 3931–3932. https://doi.org/10.1093/bioinformatics/bti630 (2005). (number: 20).
Elofsson, A. et al. Methods for estimation of model accuracy in CASP12. Proteins Struct. Funct. Bioinform. 86, 361–373. https://doi.org/10.1002/prot.25395 (2018) (number S1).
Samudrala, R. & Moult, J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 275, 895–916. https://doi.org/10.1006/jmbi.1997.1479 (1998) (number: 5).
Summa, C. M. & Levitt, M. Near-native structure refinement using in vacuo energy minimization. Proc. Natl. Acad. Sci. 104, 3177–3182. https://doi.org/10.1073/pnas.0611593104 (2007) (number 9).
Amir, E.-A. D., Kalisman, N. & Keasar, C. Differentiable, multi-dimensional, knowledge-based energy terms for torsion angle probabilities and propensities. Proteins Struct. Funct. Bioinform. 72, 62–73. https://doi.org/10.1002/prot.21896 (2008) (number: 1).
Levy-Moonshine, A., Amir, E.-A. D. & Keasar, C. Enhancement of beta-sheet assembly by cooperative hydrogen bonds potential. Bioinformatics 25, 2639–2645. https://doi.org/10.1093/bioinformatics/btp449 (2009). (number: 20).
Cheng, J., Randall, A. Z., Sweredoski, M. J. & Baldi, P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 33, W72–W76. https://doi.org/10.1093/nar/gki396 (2005).
Wang, S., Li, W., Liu, S. & Xu, J. RaptorX-Property: A web server for protein structure property prediction. Nucleic Acids Res. 44, W430–W435. https://doi.org/10.1093/nar/gkw306 (2016). (number: W1).
McGuffin, L. J., Bryson, K. & Jones, D. T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405, https://doi.org/10.1093/bioinformatics/16.4.404 (2000). (number: 4).
Sidi, T. & Keasar, C. Redundancy-weighting the pdb for detailed secondary structure prediction using deep-learning models. Bioinformatics (2020).
Zhou, H. & Skolnick, J. GOAP: A generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys. J. 101, 2043–2052. https://doi.org/10.1016/j.bpj.2011.09.012 (2011). (number: 8).
Ke, G. et al. LightGBM: A highly efficient gradient boosting decision tree. in Advances in Neural Information Processing Systems (Guyon, I. et al. eds.). Vol. 30. 3146–3154. (Curran Associates, Inc., 2017).
Pedregosa, F. et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Chen, T., He, T., Benesty, M., Khotilovich, V. & Tang, Y. Xgboost: Extreme Gradient Boosting. R Package Version 0.4-2 1-4 (2015).
Kullback, S. Information Theory and Statistics (Courier Corporation, 1997).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637. https://doi.org/10.1002/bip.360221211 (1983). (number: 12).
Wang, S., Weng, S., Ma, J. & Tang, Q. DeepCNF-D: Predicting protein order/disorder regions by weighted deep convolutional neural fields. Int. J. Mol. Sci. 16, 17315–17330. https://doi.org/10.3390/ijms160817315 (2015). (number: 8).
Levy-Moonshine, A., Amir, E.-A.D. & Keasar, C. Enhancement of beta-sheet assembly by cooperative hydrogen bonds potential. Bioinformatics 25, 2639–2645 (2009).
Pettersen, E. F. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. PubMed 30, 70–82. https://doi.org/10.1002/pro.3943 (2021).
Alvarado, S. K., Wang, Z., Miller, M. D., Thorson, J. S. & Phillips Jr, G. N. Atmm with bound rebeccamycin analogue. https://www.rcsb.org/structure/6uv6 (2020).
Alvarado, S. K., Wang, Z., Miller, M. D., Thorson, J. S. & Phillips Jr, G. N. Crystal structure of apo atmm. https://www.rcsb.org/structure/6uvq (2020).
Chen, X. et al. Protein model accuracy estimation empowered by deep learning and inter-residue distance prediction in casp14. Sci. Rep. 11, 1–12 (2021).
Kryshtafovych, A. et al. Assessment of the assessment: Evaluation of the model quality estimates in casp10. Proteins Struct. Funct. Bioinform. 82, 112–126 (2014).
Wang, W., Wang, J., Li, Z., Xu, D. & Shang, Y. Mufoldqa_g: High-accuracy protein model qa via retraining and transformation. Comput. Struct. Biotechnol. J. 19, 6282–6290 (2021).
McGuffin, L. J. & Roche, D. B. Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics 26, 182–188 (2010).
Ye, L. et al. Improved estimation of model quality using predicted inter-residue distance. Bioinformatics 37, 3752–3759 (2021).
Hou, J., Wu, T., Cao, R. & Cheng, J. Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13. Proteins Struct. Funct. Bioinform. 87, 1165–1178. https://doi.org/10.1002/prot.25697. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.25697 (2019).
Kosloff, M. & Kolodny, R. Sequence-similar, structure-dissimilar protein pairs in the pdb. Proteins Struct. Funct. Bioinform. 71, 891–902. https://doi.org/10.1002/prot.21770. https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.21770 (2008).
Sidi, T. & Keasar, C. Redundancy-weighting the PDB for detailed secondary structure prediction using deep-learning models. Bioinformatics 36, 3733–3738. https://doi.org/10.1093/bioinformatics/btaa196 (2020).
Yanover, C., Vanetik, N., Levitt, M., Kolodny, R. & Keasar, C. Redundancy-weighting for better inference of protein structural features. Bioinformatics 30, 2295–2301. https://doi.org/10.1093/bioinformatics/btu242 (2014).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science (American Association for the Advancement of Science)https://doi.org/10.1126/science.abj8754 (2021).
Acknowledgements
The authors are grateful to Tomer Sidi for creating the benchmark used in this study, to Ilya Kaufman for fruitful discussions, and to Ofir Ezrielev for the careful reviewing.
Author information
Authors and Affiliations
Contributions
C.K. conceived this study, supervised it, and curated the training data. M.B designed the experiments, performed them, and collected and analyzed the results. The manuscript was jointly written by both authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bitton, M., Keasar, C. Estimation of model accuracy by a unique set of features and tree-based regressor. Sci Rep 12, 14074 (2022). https://doi.org/10.1038/s41598-022-17097-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-17097-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.