Abstract
Drug Discovery is an active research area that demands great investments and generates low returns due to its inherent complexity and great costs. To identify potential therapeutic candidates more effectively, we propose protein–ligand with adversarial augmentations network (PLA-Net), a deep learning-based approach to predict target–ligand interactions. PLA-Net consists of a two-module deep graph convolutional network that considers ligands’ and targets’ most relevant chemical information, successfully combining them to find their binding capability. Moreover, we generate adversarial data augmentations that preserve relevant biological backgrounds and improve the interpretability of our model, highlighting the relevant substructures of the ligands reported to interact with the protein targets. Our experiments demonstrate that the joint ligand–target information and the adversarial augmentations significantly increase the interaction prediction performance. PLA-Net achieves 86.52% in mean average precision for 102 target proteins with perfect performance for 30 of them, in a curated version of actives as decoys dataset. Lastly, we accurately predict pharmacologically-relevant molecules when screening the ligands of ChEMBL and drug repurposing Hub datasets with the perfect-scoring targets.
Similar content being viewed by others
Introduction
The development of novel drugs with therapeutic potential is a challenging yet essential endeavor to ensure human welfare and actively confront health threats. This process is characterized by conventional cycles lasting up to 12 years and demanding costs of about 2.7 billion US dollars1, with declining compensation due to limited efficacy and safety issues during clinical trial stages2. In this regard, the likelihood of approval for small molecule candidates for the period 2011–2020 was only 7.5%3, with expenditures ranging from 800 million to 1.4 billion US dollars in unsuccessful clinical trials4. These large yet unfruitful investments are currently forcing pharmaceutical and related sectors to search for more efficient strategies in terms of research objectives and profitability.
To address these shortcomings, reverse pharmacology, also known as target-based screening, proposes more robust data-driven approaches to improve the identification of active molecules towards therapeutic biological targets5. This methodology starts by detecting malfunctioning proteins as therapeutic targets of specific diseases through animal models or interdisciplinary analysis of patients phenotype and genotype. Once the targets are selected, high-throughput screenings are performed to detect the best pharmacological candidates against those targets6. This approach’s main promise is the possibility to increase predictability and accuracy of drug screening processes by systematizing analyses over large pharmaceutical datasets, which results in lessening the need for laborious in vitro and in vivo experimentation, and a significant increase in the overall efficiency of the entire discovery process. In this regard, virtual screening emerges as the current standard for the prediction of interactions between small molecules and proteins7. Conventional methods in virtual screening mainly employ molecular docking techniques, but are still limited in accuracy and effectiveness, and often require expensive experimental testing for validation prior to market launching8,9. In consequence, there is still a large scope for improving screening processes, especially regarding biological compatibility prediction.
Recent works use deep learning (DL) techniques in broad applications of the molecular biology domain, whose understanding is critical for the development of medicine and the comprehension of the biological interactions during physiological processes. For example, the prediction of functions for programmable RNA switches10,11, the three-dimensional structure prediction of proteins12,13, the prediction between protein-protein interfaces14, and the discovery of structurally distinct novel antibiotics15 are tasks in which DL has enabled the extraction of useful data and helped reduce laboratory experimentation. As in the research topics mentioned above, the interaction between small molecules and proteins is essential in the development of medicine, and therefore, human welfare. The information provided by the analyses of these interactions, such as the functional units, the binding pockets, and interaction sites between drugs and targets, is crucial for the targeted development and design of pharmaceuticals. For this reason, it is of general interest to know both if there is a target–ligand interaction and how this interaction occurs.
In this perspective, DL methods might be able to play a pivotal role due to their ability to find and exploit patterns in large datasets and distill salient features that characterize effective biological interactions between cellular targets and small molecules, henceforth regarded as target–ligand interactions (TLIs)16. Therefore, we address one of the most common formulations for TLI understanding: the binary classification of active and non-active interactions between a ligand and a protein. This task substantiates the larger endeavor of the niche market for drug discovery and repurposing, whose size is projected to grow from 24.96 to 34.62 billion US dollars between 2020 and 202717.
Understanding biological interactions of pharmacological molecules involves reasoning over their intricate structures and corresponding traits at the atomic level. This working objective stresses the importance of appropriate molecular structure representation. In this respect, previous approaches have employed two-dimensional molecular images built from one-hot embeddings over atoms, a general method to vectorize categorical features, and have been analyzed under convolutional neural networks (CNN)18 and recurrent neural networks (RNN)19 to predict TLIs. However, these contributions disregard molecular structure, as well as the target protein’s information. Alternatively20 and21 incorporate the 3-dimensional (3D) structural information of the ligand positioned in the target receptor to predict activity through CNNs. Nonetheless, this type of input complex needs additional pre-processing and previous knowledge of the 3D structure of the protein, its binding pockets and the ligand position within the pocket, which are not easily available. Given the non-Euclidean nature of chemical data, molecular graph representations enable a more accurate and explicit modeling of atom and bond spatial configurations. Moreover, the conjunction of graph representation and deep learning techniques provides a promising approach to model molecular structures, both of small molecules and proteins, while extracting features with relevant biological backgrounds22,23.
Despite recent success in improving TLI predictions, their interpretability is a recurrent issue to validate their translation into compelling medicine scenarios. This feature is particularly important since high-performance metrics fail to provide sufficient information for evaluating if the model is learning relevant biological and chemical data for pharmacological design. To circumvent this situation, attention methods have emerged as powerful tools to identify key substructures in both targets and ligands for the model’s predictions23. Zheng et al., for instance, implemented a multi-head attention algorithm for both target and ligand embeddings to visualize more easily where the model focuses to predict TLIs24. Although their method is able to identify real overlapping regions of interaction in both ligands and targets, they disregard structural information by using linear representations of the molecules as input.
In this work, we propose the application of graph convolutional networks (GCNs) for predicting TLIs, where graph-based representations of both molecules and target proteins are obtained from easily accessible simplified molecular-input line-entry system (SMILES) strings and amino acid sequences in FASTA format. We address the most common limitations of GCNs, such as depth limits due to vanishing gradients, over-smoothing and loss of spatial information, by building on the work of25 and26 in the context of TLIs. We harness their proposed method for extrapolating common CNN strategies (e.g., residual and dense connections, dilated convolutions) to GCNs, which they demonstrated capable of enabling the training of deeper networks for a variety of computer vision tasks. This contrasts with previous graph-based methods proposed for molecular property prediction where limited depth and small receptive fields are among their main limitations. For instance, DeepChem equipped with graph convolutional model from27, and ChemProp15 have up to six message-passing layers, and PotentialNet28,29 has 3 stages of graph spatial convolution. Accordingly, we expect that employing deeper networks with larger receptive fields will favor the learning of global chemical information and will be reflected on overall performance. Moreover, we propose a method for increasing the interpretability of our TLI predictions by training the networks with adversarially-augmented molecules. These, in turn, are inspired by adversarial examples, specialized inputs with small intentional feature perturbations that cause machine learning models to make false predictions30. These have gained increasing popularity over the past few years in the field of computer vision due to their ability to direct learning towards semantically-aligned features31,32. In particular, we introduce a biologically-bounded gradient-based method to generate adversarial molecule augmentations, which adapts gradient-based edge deletion methods previously proposed for adversarial examples in graph data33 while preserving biological consistency and essential class features in molecular graphs.
Our network, henceforth termed protein–ligand with adversarial augmentations network (PLA-Net), comprises two modules that separately extract information from proteins and ligands, thereby learning optimal representations for further compatibility analysis (Fig. 1). Our contributions are three-fold: (1) we model the spatial configuration of both the target protein and the ligand through bidirected graphs, (2) we integrate relevant chemical/primary structure information from proteins for TLI prediction, and (3) we propose a gradient-based method to compute adversarial molecule augmentations that preserve relevant biological backgrounds and improve both interpretability and overall performance. We train PLA-Net models for 102 pharmacologically-relevant protein targets and establish the new state-of-the-art, outperforming the previous one19 by a large margin of 19.8% on mean average precision (mAP) in a curated version of the actives as decoys (AD) dataset. Moreover, we perform a virtual screening between molecules of two large datasets (ChEMBL34, and Drug Repurposing Hub35) with the perfect-scoring targets, and corroborate that our method accurately predicts experimentally validated TLIs. We also demonstrate that PLA-Net is able to identify relevant substructures in molecules reported to interact with proteins of clinical interest. Both the outstanding performance and the interpretable results position PLA-Net as a promising starting point to integrate deep learning methods into drug discovery pipelines.
PLA-Net workflow. Schematic representation of a PLA-Net model for predicting interactions between small organic molecules and one of the 102 target proteins in the AD dataset. Graph representations of the molecule and a given target protein are generated from SMILES and FASTA sequences and are used as input to the Ligand Module (LM) and Protein Module (PM), respectively. Each module comprises a deep GCN followed by an average pooling layer, which extracts relevant features of their corresponding input graph. Both representations are finally concatenated and combined through a fully connected layer to predict the target–ligand interaction probability. Created with BioRender.com.
Results and discussion
PLA-Net considerably outperforms state-of-the-art models in the proposed benchmark
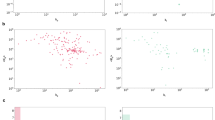
Figure 2a shows that PLA-Net significantly improves TLI prediction and outperforms by a large margin TLI state-of-the-art methods19,36 and DeeperGCN26 trained for this task. Besides increasing over 19 points in mAP from the highest-performing method19, the performance distribution of PLA-Net is superior than in the referred methods. Figure 2b shows that PLA-Net considerably shifts the performance histogram of the 102 models to the right, when compared to the current state-of-the-art method (PharmaNet)19. The models for 47 targets in PLA-Net achieve performances above 95% AP, which contrasts with only 18 targets in the case of PharmaNet. Moreover, 30 of these models achieve perfect scores, compared to only 13 in PharmaNet19. This suggests that the method and training curriculum proposed may enable a more explicit modeling of TLIs than previous works. The performance of all PLA-Net models is listed in Supplementary Table 1.
Other descriptors such as number of amino acids (AAs), intraclass similarity and number of actives/decoys per target are also included for comparison. However, we found no apparent correlation between these descriptors and the performance of PLA-Net models. The size of protein targets varied between 100 and 1434 AAs, with large and small proteins having both high and low performances. For instance, NOS1 (1434 AAs) and IGF1R (1367 AAs) achieve performances of 98.89% and 23.31%, while PA2GA (144 AAs) and TRY1 (247 AAs) achieve performances of 100% and 50.02%, respectively. Most importantly, within the 30 perfect-scoring targets there were both small proteins, such as FKB1A (108 AAs) and PA2GA (144 AAs), and large proteins, such as ROCK1 (1324 AAs) and ITAL (1170 AAs). Likewise, high and low performing protein models exhibit similar intraclass similarity distributions, which vary between 0.78 and 0.93. This suggests that performance differences may be merely a result of varying difficulties between TLI tasks.
Comparison with state-of-the-art methods trained for TLI prediction in the proposed benchmark. (a) Performance distribution curves comparing our model (PLA-Net) with state-of-the-art methods. For each model, we show the number of binary models that achieve a TLI prediction performance greater than or equal to a specific AP value. (b) We compare the performance distribution of the 102 targets in PLA-Net with that of the current state-of-the-art (PharmaNet)19, showing that PLA-Net consistently improves the AP metric of the majority of the targets, with 59 targets with performance between 90 and 100% versus 29 in PharmaNet. Furthermore, PLA-Net achieves perfect performance for 30 targets with high clinical interest.
Graph representations capture more relevant molecular information than linear representations
To exploit the information-rich graph representations proposed, we optimized the parameters of our LM (e.g., optimizer, scheduler, average pooling method, depth) to boost its discriminative power. Ablations were performed over 15 representative targets, selected according to varying levels of difficulty are shown in Table 1. We compared the performance of the optimized LM with a classical machine learning technique (i.e., a random forest), and the current state-of-the-art approach (i.e., PharmaNet)19. The random forest takes as input a 6027 feature vector, extracted following37’s definition of atom pair descriptors, while PharmaNet takes as input the SMILES strings for each molecule. Since these methods were originally trained in a multiclass setup, we trained our LM in the same setting for fair comparisons. Additionally, we retrained PharmaNet in our proposed binary classification setup to compare their performance in both cases. We ablated the protein information from our model to ensure a fair comparison between methods.
Table 2 shows that our LM outperforms the random forest classifier and PharmaNet models in both multiclass and binary setups. In particular, the LM trained in the binary setup achieves performances over 5% and 10% higher than these competing models. These results highlight the importance of structural information granted by graph representations of ligands, especially when training binary models for predicting TLIs with each target. Even though training one model is inherently more efficient, this training time is counteracted by the benefit of models achieving highly accurate predictions for specific therapeutic targets with clinical relevance. Accordingly, all experiments were performed in the binary setup.
Including protein information improves TLI predictions.
Since the information of target proteins is fundamental when assessing a TLI, we hypothesized that its inclusion would enhance the classifier’s discriminative power between actives and decoys. In particular, we expected that training the PM jointly with the pre-trained LM would promote the former to learn features specifically relevant to the target protein’s interaction with its ligands. As expected, including protein information increases the performance of 66.7% of the 102 models and improves the mAP metric by 1.19 points when compared to the LM alone (Fig. 3). The incorporation of target protein information during LM + PM training was optimized by including a learnable parameter that multiplied the fully connected layer weights associated with the protein information, thus controlling their contribution. We also show that initializing this multiplier as zero increases the performance of our models by allowing a smoother incorporation of this data when compared to random initializations (Fig. 4). This ablation was performed over the models for the same 15 representative targets selected above and resulted in an increase of 7.08 points in the mAP metric with respect to randomly initialized weights.
To verify that incorporating protein information is effectively improving the predictive power of our model, instead of just prolonging training, we also trained the 102 models replacing the output of the PM by a constant 128-D vector of ones. This ablation resulted in an average performance of 82.45% mAP, which is slightly lower than when training the LM alone. This was an expected result considering that the vector of ones is not contributing additional information about the TLI, which forces the model to focus mainly on the information provided by the LM. Moreover, it corroborates that the learned representations by the PM are contributing valuable information for the TLI prediction.
Performance distribution of PLA-Net training stages. The performance of individual targets shows a marked tendency towards high and perfect mAP scores (90–100%) as the training curriculum progresses. In particular, LM \(+\) PM and augmented LM (LM \(+\) A) show a clear improvement in performance distribution with respect to LM, and this is further improved when combining the information extracted by each in PLA-Net. Best viewed in color.
Initialization of protein contribution during LM \(+\) PM training. Zeroing the linear classifier’s weights that correspond to the protein contribution at the onset of the training (I0) substantially improves the performance compared to a random initialization of the protein contribution (RI). We measured performance in mAP for 15 representative targets.
Adversarial data augmentations improve model performance and interpretability
We propose the inclusion of adversarial molecule augmentations specifically tailored to harness the model’s weaknesses during training. Accordingly, we intend to help the model learn semantically-aligned features of the active molecules for each target and effectively discriminate them from decoys. Following the augmentation process described in “Adversarial data augmentations improve model performance and interpretability” section, we generated adversarial examples of the active molecules to each target at each training stage. This augmentation regime improved the average performance of the LMs alone by 3.25 points, increasing the performance of 55% of the 102 models and maintaining the performance of 40% (Fig. 3). In particular, 13 of the models increased by more than 10 points in AP. Supplementary Fig. 1 shows that the LM and the augmented LM of the 15 representative targets described above converge early in training, which verifies that the observed increase in performance was not merely due to the increase in computation. Overall, this suggests that the generated active molecules in each training stage were sufficiently similar to experimentally-proven ligands to maintain class consistency but also sufficiently different for the model to learn relevant features that it failed to learn with the original dataset.
To assess the effect of the proposed training schemes over the interpretability of our model, we conducted a gradient-based analysis to highlight the learned molecular salient features in each scenario. The atoms with the lowest gradient within the molecular graphs of active molecules were interpreted as the most important for the TLI prediction, due to their highest contribution towards minimizing the loss function. To exemplify this analysis, Fig. 5 compares the predicted importance of ligand substructures when training with augmented molecules (LM \(+\) A), with protein information (LM \(+\) PM) or with neither (LM). We also show substructures that have been previously reported to participate in the TLI of that specific ligand with its target to validate the models’ predictions. Notably, LM \(+\) A and LM \(+\) PM exhibit marked attention shifts towards localized substructures that have been previously reported to be involved in the respective TLIs. For instance, adenine and ribose substructures are predicted to be the most important for TLIs with the adenosine A2 receptor (AA2AR) by LM \(+\) A and LM \(+\) PM models, respectively, which coincides with the importance of adenosine substructures commonly found in AA2AR ligands39. In contrast, the pristine LM model fails to capture this information despite achieving an AP only 1% lower. Similarly, the glutamate backbone in ligands for the glutamate ionotropic receptor (GRIK1) is partially or completely highlighted when employing either of these training schemes, while it is given the least importance when training without them (LM). Considering that the pristine LM model for TLI prediction with GRIK1 already achieves a perfect-score, this suggests that the additional information included during LM \(+\) A and LM \(+\) PM training is shifting model training towards learning interpretable features beyond improving performance. This statement also holds true for other targets such as the adenosine deaminase (ADA), dihydrofolate reductase (DYR) and the epidermal growth factor receptor (EGFR), although much more prominently for adversarially-augmented models. In these cases, LM \(+\) A is able to focus on relevant substructures whose interaction with the target’s binding pocket has been previously described with molecular docking analyses (e.g., azole groups for ADA40, 4-amino groups and pteridine rings for DYR41 and quinilone N1 and nitrile groups for EGFR42). Moreover, the importance of specific functional groups directly involved in the function of enzymatic targets is also best captured by LM \(+\) A models, such as carbonyl groups in the ligands for 11-b-hydroxysteroid dehydrogenase 1 (DHI1), which are reduced into hydroxyl groups upon their interaction with DHI143.
However, the attention shifts induced by the inclusion of target protein information in some LM \(+\) PM models was not as fruitful as when training with augmented molecules. Phenol groups in ligands for the estrogen receptor alfa (ESR1)44, for example, lose importance in LM \(+\) PM models despite being directly implicated in the TLI of this ligand with its receptor. Moreover, the importance of the N2 and nitrile groups of quinilone42 is completely lost when training with the protein information of EGFR, even though the AP achieved by the LM \(+\) PM model for this target is higher than that of the LM model. Overall, these results suggest that the observed increase in interpretability is not merely a result of the increase in model performance, but that the type of additional information does play a role in what the models are learning. In particular, the additional information provided by the generated adversarial molecules appears to be the most useful in directing training towards the learning of semantically-aligned features, which is one of the major challenges in the training of generalizable neural networks45.
Salient feature maps of ligands during PLA-Net training stages. Salient feature maps predicted by the LM trained only on original molecules (LM), the LM trained with adversarial augmentations (LM \(+\) A), and the LM and PM jointly trained (LM \(+\) PM) for representative ligands of 7 protein targets. The average precision (AP) of each model is presented below their respective feature map and TLI-relevant substructures are shown to the left. All of these substructures have been previously identified through experimental and/or molecular docking analyses between the shown ligand and its respective target protein39,40,41,42,43,44. The predicted importance of ligand substructures significantly shifts at each training stage despite small changes in AP. The augmented LM achieves predictions that best align with substructures of natural ligands that have been previously reported to participate in TLIs. Created with BioRender.com.
PLA-Net’s perfect scoring models predict pharmacologically-relevant TLIs on drug repurposing and ChEMBL databases
To validate that PLA-Net models were able to make reasonable predictions in previously unseen data, we perfomed a high-throughput virtual screening for pharmacologically-relevant molecules on Drug Repurposing and ChEMBL databases. In particular, we predicted TLIs with 9 protein targets that achieved perfect scores during training and are closely related to different pharmacological applications (Table 3). We validated the top predictions of our models by manually corroborating them with previous literature reports. Figure 6 shows five molecules from each database that PLA-Net predicted to interact with each target with a high probability. Red scores correspond to the Rogot–Goldberg similarity between the predicted ligands and the active molecules in the training set. Green labeled molecules have been previously reported as active for the target of interest, yellow labeled molecules have been reported as active for a closely related protein, and orange labeled molecules have not been reported but exhibit relevant substructures present in active molecules for the same target.
PLA-Net’s pharmacologically-relevant TLI predictions on the Drug Repurposing and CHEMBL databases. From each database, were selected five molecules predicted as active with high probability for nine pharmacologically-relevant targets. The name and prediction probability for each molecule are shown in their upper right corner. The mean Rogot–Goldberg similarity between each molecule and the active molecules of the corresponding training set is shown in red in their lower right corner. The mean Rogot–Goldberg similarity between each molecule and the active molecules of the corresponding training set is shown in red in their lower right corner. Molecules’ activity towards each target was corroborated with previous literature reports57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118 . Green label: experimentally-proven active molecule for the respective target. Yellow label: experimentally-proven active molecule for protein closely related to the target of interest. Orange label: not experimentally-proven, but with relevant substructures present in experimentally-proven active molecules for the target. Created with BioRender.com.
PLA-Net was able to accurately predict, for all target proteins, multiple molecules that have been previously reported as active towards each target, as well as molecules that follow consistent structural patterns. All molecules predicted as active for farnesyl pyrophosphate synthase (FPPS), for example, include biphosphate groups within their structure, which is a recurrent hallmark of FPPS ligands given the active participation of this group in their interaction with FPPS46. Accordingly, all shown predictions have been previously reported as FPPS inhibitors. Moreover, the relatively low fingerprint similarities between the predicted ligands and the training set ligands (0.67-0.71) suggest that, despite the consistent appearance of biphosphate groups, side-chain elements can vary widely. This demonstrates that the model is capable of prioritizing the presence of biphosphate groups and ignoring side-chain differences. Similarly, most molecules predicted as ligands towards PNPH, the purine nucleoside phosphatase, contain a purine substructure that correlates well with the enzyme’s function. In turn, all molecules whose interaction has not been reported are either substructures of reported molecules or have slight ramification changes of known active molecules. This pattern was consistently observed with other targets such as the glucocorticoid receptor (GCR), the thymidine kinase (KITH) and the adenosylhomocysteinase (SAHH). In these cases, steroid backbones, thymine substructures and adenine substructures were present in most predicted ligands, respectively. Notably, despite these consistent structural patterns, many experimentally-validated predictions for these targets also exhibit low similarities with active training molecules (0.56–0.59), which again suggests that these models are capable of recognizing consistent substructures within heterogenous backbones.
In contrast, some predicted molecules for targets such as SAHH, adenosine deaminase (ADA) and phospholipase A2 (PA2GA) showed no clear structural pattern, yet they still fit into one of two categories: (1) have been reported as active towards the protein or closely related proteins, or (ii) are substructures of active molecules for the target protein. In the particular case of SAHH, the predicted molecule Inositol has been experimentally proven as active towards this protein47, but its structure largely differs from the other experimentally validated predictions and active ligands from the training set (0.5602 average fingerprint similarity). Similarly, the predicted active molecule for PA2GA, 1-hexadecanol, is a substructure of its experimentally-proven ligand 2-Ethylamino-1-hexadecanol48, which fails to exhibit the purine-based structures observed previously in other experimentally-proven ligands for this target. The same trend is observed with the alcohol derivatives predicted for ADA, which are substructures of other previously reported ligands49,50 and their presence has shown to favor TLI occurrence51. This suggests that our model is able to learn relevant substructures from heterogeneous molecule sets and is not merely memorizing single structural patterns. Further evidence of this finding can be found by observing that several experimentally proven ligands for the C-X-C chemokine receptor type 4 (CXCR4) and the glutamate ionotropic receptor kianate 1 (Grik1) were accurately predicted, despite their marked structural differences and no apparent common backbone (0.52–0.63 fingerprint similarity with training sets). The observed discriminative power correlates well with the marked heterogeneity in the training sets used for these two targets, which comprise a wide array of molecules with divergent structural backbones and limited congruencies. For instance, although all Grik1 ligands coincide in a terminal glutamate-derived functional group, the large backbones differences between them are well captured within the dataset (Supplementary Fig. 2). Accordingly, although some predicted ligands are closely respresented by active molecules in the training set (e.g., L-quiscalic acid, CHEMBL4066795), the others that are not can still be accurately identified (e.g., CHEMBL199233, tetrazol-5-yl-glycine, CHEMBL283725). Furthermore, PLA-Net’s prediction scores are significantly higher than the similarity score, highlighting the advantage of our method over canonical fingerprint analysis. Overall, these highly accurate predictions validate that PLA-Net is effectively learning underlying structural hallmarks that dictate TLIs with protein targets. This is of special clinical relevance given their close involvement in numerous diseases such as cancer, chronic inflammation, neurodegeneration, and microbial infections.
Conclusion
In this work, we propose PLA-Net, a two-module deep GCN to tackle TLI prediction in a curated version of the AD dataset. Our method merges rich information extracted from ligand and protein graphs utilizing deep GCN modules. Additionally, we propose a method for generating adversarial molecule augmentations that preserve biologically relevant backgrounds and show that their inclusion during training improves our model’s performance and interpretability. Accordingly, PLA-Net not only becomes the new state-of-the-art in TLI prediction, but allows a more comprehensive analysis of the underlying features dictating TLIs. Moreover, its highly accurate TLI predictions with molecules unseen previously by the network and extracted from large and unannotated databases brings us one step closer towards interpretable pharmaceutical discovery.
Methods
TLI benchmark
The Database of Useful Decoys Enhanced (DUD-E) is widely employed to benchmark approaches that predict TLIs52,53,54. DUD-E contains 22,886 experimentally verified active compounds towards 102 proteins of clinical interest listed in Supplementary Table 1. Some of these proteins are related to chronic diseases of significant clinical interest such as hypertension (RENI), HIV (CXCR4), cancer (FPPS), and Parkinson’s disease (COMT). Additionally, for each active molecule, DUD-E contains 50 non-active compounds (decoys) with similar physicochemical properties but different topologies. This unbalanced data distribution replicates the real scenario of finding an active compound against a huge variety of decoys.
However, Chen et al.36 proved that DUD-E’s selection criteria for the decoys follows a pattern that makes them easily distinguishable from the active molecules. Conversely, they proposed the Actives as Decoys (AD) dataset, in which the decoys are selected from the active molecules of other protein targets. In particular, the decoys for a protein are selected by performing molecular docking between that protein and the active compounds from the other 101 proteins. The TLI is then ranked based on the predicted binding energy. The top-50 molecules of each of the 101 targets are selected to be the decoys of that target protein. This process is repeated for all targets to create the decoy dataset.
Nonetheless, we noticed that some molecules in the DUD-E dataset are labeled as active for more than one protein target and, therefore, the decoy selection process of the AD dataset causes some molecules to be simultaneously labeled as active and decoy for the same protein. In addition, most decoys for the same protein are repeated. Since this contradictory and redundant information might detrimentally lead to a bias during model training, we removed the repeated samples (546,412 molecules) and variably labeled samples (37 molecules) for each target, which represent 52.29% of the original data. We also ensure that active compounds of the targets are the same as those experimentally validated in DUD-E. Subsequently, we separated the curated version of the AD dataset into training and testing sets, comprised of 90% and 10% of the compounds for each protein, respectively. Next, we performed a four-fold cross-validation with the training subset. Protein information for the 102 protein targets in the AD dataset was obtained by compiling their FASTA sequences from the Universal Protein (UniProt) repository.
We evaluated with the Average Precision (AP) metric and report the mean AP (mAP) of the predicted interactions with the 102 target proteins.
Molecule representation
The molecular representation used by state-of-the-art methods in the AD Dataset, SMILES, fails to recapitulate the complexity of molecules mainly due to the absence of structural information that dictates interactions between atoms. Other methods rely on very complex 3D representations, which cannot be obtained for all protein-ligand pairs because they depend on experimental techniques such as X-ray crystallography and computational techniques such as molecular docking. In contrast, graph representations are well suited for capturing molecular information since they enable the reconstruction of atomic networks that preserve chemical and structural information. For this reason, we built the graph representations of both ligands and target proteins based on their atomic and bond configuration. The RDKit package was used to convert their respective SMILES and amino acid sequences in FASTA format into molecular graph representations (Fig. 1). Given a ligand or protein, its graph is represented as \(\mathscr {G} = (\mathscr {V},\mathscr {E},X_v,X_e)\), where \(\mathscr {V}\) denotes the set of atoms (nodes), \(\mathscr {E}\) the set of bonds (edges), \(X_v\) the set of atom features and \(X_e\) the set of bond features. An atom feature vector \(x \in X_v\) comprises nine properties of the atom \(v \in \mathscr {V}\): atomic number, chirality, degree, formal charge, number of hydrogens, number of radical electrons, hybridization, aromaticity and ring membership. Similarly, we built a bond feature vector \(x_{vu} \in X_e\) from three characteristics of the bond \(e_{vu} \in \mathscr {E}\) between atom v and atom u: type of bond, stereochemistry, and conjugation. Each feature is represented as a one-hot vector and all one-hot feature representations are then concatenated to form the feature vector. The length of each one-hot vector is determined by the possible options for describing each feature, which are summarized in Table 4. Moreover, all bonds are assumed bidirectional, \(X_{e_{vu}} = X_{e_{uv}}\). Accordingly, this graph representation enables the learning of both chemical and spatial distribution properties due to the node/edge feature vectors and edge configurations between nodes, respectively. Furthermore, this representation favors the learning of implicit 3D information that is inherent to the modeled molecules such as the stereochemistry of bonds, chirality, and aromaticity. Moreover, note that geometrically optimized 3D models of either ligands or proteins are not necessary for graph construction due to the ease of extracting all necessary information directly from SMILES and FASTA strings, making this process computationally inexpensive.
Architecture details
As shown in Fig. 1, ligand and protein graphs are analyzed separately by a GCN module, which extracts chemical and spatial information to obtain optimal representations for further TLI analysis. Each module comprises a deep GCN and an average pooling function, which outputs a 128-D vector representation for the molecular graph. Both optimized representations are concatenated, a linear layer merges the information, and a fully connected layer then classifies the ligand as an active or decoy.
Our GCN modules were adapted from26, which is a message-passing framework originally designed for molecular property prediction. Being \(\mathscr {N}_v\) the set of neighbors of atom v, the message passing algorithm of the lth layer is described by message construction (Eq. 1), message aggregation (Eq. 2) and node update (Eq. 3) functions.
where \(\rho ^{(l)}\), \(\zeta ^{(l)}\), \(\phi ^{(l)}\) are all learnable and differentiable functions. The message construction function \(\rho ^{(l)}\) is applied to the features of atom v, the features of its neighbor u, and their corresponding edge features \(x_{vu}\) to obtain an individual message \(\mathscr {M}_{vu}^{(l)}\) for each neighbor \(u \in \mathscr {N}_v\). The message aggregation function \(\zeta ^{(l)}\) takes as input the set of individual messages and outputs an aggregated message. In this case, \(\zeta ^{(l)}\) is a learnable softmax function. Finally, \(\phi ^{(l)}\) updates the node features of the l-th layer by adding the aggregated message \(\mathscr {M}_{v}^{(l)}\) and passing it through a multi-layer perceptron.
As mentioned in25 and26, to enable the adequate training of deep GCNs this architecture employs two techniques inspired in CNNs: (1) pre-activation variants of residual connections, and (2) dilated aggregations that enlarge the receptive field. In the former, training proceeds by a change in the usual ordering of GCN components. In other words, instead of performing the graph convolution, followed by normalization, a ReLU layer and the addition of the residual connection, this architecture performs first the normalization and ReLU activation, followed by the graph convolution and the addition of the residual connection. In the latter, inspired by dilated convolutions in CNNs, a dilated graph is constructed after each message passing layer55. In particular, the network relies on a Dilated k-NN to find dilated neighbors at a d dilation rate. This operation returns the k nearest neighbors within the kxd neighborhood region by skipping every d neighbors.
Our modules consist of 20 message-passing layers, and final embedding size for nodes’ and edges’ features of 128.
Adversarial augmentations. (a) Augmented molecules are generated through an edge-deletion process that selects the edge of the molecular graph to delete by following two criteria: (i) the deletion of the selected edge must generate an adversarial molecule whose distance to the Bemis–Murcko scaffold of the original molecule is less than a defined threshold (\(\mu\)) and (ii) the selected edge must have a negative gradient and the gradient magnitude must be maximal. (b) Comparison of intra-class distance as a function of different similarity metrics. We computed the distances between the Morgan fingerprints of molecules from a specific target class and of their corresponding Bemis–Murcko scaffolds to assess the average intra-class distance as a function of different similarity metrics. We selected the Rogot–Goldberg similarity descriptor due to its high performance for intra-class similarities. Created with BioRender.com.
Adversarial data augmentations
To generate biologically relevant adversarial molecules, we propose a gradient-based method that modifies the edges of molecular graphs according to their contribution to the model’s outcome. In particular, we associate a binary coefficient with a value of 1 to each edge in the graph and multiply it with the feature vector of its corresponding edge during graph construction. Even though this multiplication maintains the original values of the feature vector and the computation of the molecule’s embeddings, the gradient of these binary variables after backpropagating through the model becomes crucial for determining which edges are contributing the most to the model’s predictions. In this context, the work by Dai et al.33 inspired us to delete the edge with the most negative gradient coefficient, that is, the one with the highest contribution in minimizing the loss function.
To ensure that the augmented molecule resulting from the deletion of the chosen edge preserves relevant class characteristics, we defined a molecular distance metric that bounds the edge selection process. We impose a distance constraint between the augmented molecule and the original molecule as a function of the similarity between the former and the Bemis–Murcko scaffold56 of the latter. This scaffold preserves crucial structural characteristics of a molecule by retaining backbone structures and eliminating side-chain elements56. We chose this scaffold as reference, instead of the original molecule, since we wanted to preserve molecular backbones in the augmented molecules. This is particularly important since modifying these structures may induce large conformational changes that could detrimentally impact the TLI under real physiological scenarios and, in turn, yield molecules that fail to preserve relevant class characteristics. Moreover, considering that Bemis-Murcko scaffolds were previously used to eliminate similar molecules while assembling the DUD-E dataset52, these scaffolds are highly suitable for molecule comparison.
The distance between augmented molecules and their respective scaffolds is computed over their Morgan fingerprints according to Eq. (4).
where \(M_s\) is the Morgan fingerprint of the Bemis–Murcko scaffold of the original molecule, \(M'\) is the Morgan fingerprint of the augmented molecule, and RGS is the Rogot–Goldberg similarity between them. RGS was chosen above other fingerprint similarity metrics (e.g., Tanimoto, Dice, Sokal, Russel) since it minimizes the average distance between the fingerprints of molecules and their corresponding Bemis–Murcko scaffolds (Fig. 7b). To bound the described distance, we define the largest distance from the molecules to their corresponding scaffolds as a threshold \(\mu\). If deletion of an edge causes the distance between the modified molecule and the scaffold of the original molecule to exceed the defined threshold, that edge is skipped to evaluate the next candidate, according to the negative gradient magnitude. If no edge with a negative gradient satisfies this condition, the molecule is skipped and no augmented molecule is retained. An example of this adversarial augmentation process is shown in Fig. 7a.
We generated augmented molecules for each batch and included them during training in addition to the original molecules. In this way, each batch included augmented molecules that exploit the strengths and weaknesses of the model at each training stage, tailoring the augmentation process to the model’s needs.
Implementation details
PLA-Net models for each of the 102 target proteins were trained following a multi-step curriculum directed towards optimizing the molecule and protein representations such that their relevant information could be easily extracted and assembled. First, the Ligand Module (LM) was individually trained with only original molecules for 300 epochs and a learning rate (LR) of 5e−3. In this case, the 128-D feature embeddings outputed by the LM is directly passed through the classification layer since no protein information is included. A randomly-initialized Protein Module (PM) was then included and jointly trained with the previously trained LM for 20 more epochs to integrate protein information relevant to the TLI. This was done following the workflow described in Fig. 1. Simultaneously, another LM was trained from scratch, with both original and augmented molecules, for 300 epochs and a LR of 5e−4. Again, the outputed 128-D feature embeddings were passed directly through the classification layer as no protein information was included. Finally, the 128-D feature embeddings generated by the PM jointly trained in the first stage, and by the augmented LM of the second stage were concatenated and their extracted information was combined with the training of a fully connected layer and classification layer for 20 epochs and a LR of 5e−5. In this final step, the concatenated information is first transformed into a 128-D vector with the fully connected layer and then passed through a classification layer that yields the binary prediction for the TLI.
Screening for drug discovery and drug repurposing
To validate the quality of our models’ predictions and propose new pharmaceutical candidates for our perfect scoring targets, we screened for TLIs on the ChEMBL (https://www.ebi.ac.uk/chembl/)34 and the Drug Repurposing Hub (https://clue.io/repurposing)35 databases. ChEMBL is a manually curated database with information for 15’504,604 bioactive molecules with drug-like properties34. Similarly, the Drug Repurposing Hub is a curated and annotated collection with 13,553 FDA-approved drugs, clinical trial drugs, and pre-clinically tested compounds developed with the aim of revealing new therapeutic targets for known drugs35.
We selected the models that achieved perfect TLI scores on the test set (i.e., models for 30 protein targets) for predicting TLIs with molecules in the Drug Repurposing Hub. Out of the selected models, we chose 11 to evaluate TLIs with ChEMBL molecules according to their clinical and therapeutic relevance. We filtered out from both databases the molecules whose SMILES could not be converted into a graph due to RDKit SMILES’-reading format. After this curation, we tested the 30 perfect-scoring models with 6,798 unique molecules of the Drug Repurposing Hub and the referred 11 models with 2’031,651 unique molecules of ChEMBL. Lastly, we selected the molecules with the 5 highest TLI scores for each tested target, after ensuring that those molecules were not considered for training the model.
Code and data availability
Code, implementation instructions, the curated AD dataset, PLA-Net weights and inference in drug repurposing and CHEMBL databases available at PLA-Net repository in https://github.com/BCV-Uniandes/PLA-Net.
References
Cui, W. et al. Discovering anti-cancer drugs via computational methods. Front. Pharmacol. 11, 72–85 (2020).
Lavecchia, A. & Cerchia, C. In silico methods to address polypharmacology: Current status, applications and future perspectives. Drug Discovery Today 21, 288–298 (2016).
Thomas, D. et al. Clinical development success rates and contributing factors 2011–2020 (2021).
Food, T. & Administration, D. Fda executive summary (2017).
Swinney, D. C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011).
Savva, K. et al. Computational drug repurposing for neurodegenerative diseases. In In Silico Drug Design 85–118. https://doi.org/10.1016/b978-0-12-816125-8.00004-3 (Elsevier, 2019).
Stanzione, F., Giangreco, I. & Cole, J. C. Use of molecular docking computational tools in drug discovery. Progress Med. Chem. 60, 273–343 (2021).
Leelananda, S. P. & Lindert, S. Computational methods in drug discovery. Beilstein J. Organic Chem. 12, 2694–2718 (2016).
Phatak, S. S., Stephan, C. C. & Cavasotto, C. N. High-throughput and in silico screenings in drug discovery. Expert Opin. Drug Discov. 4, 947–959 (2009).
Angenent-Mari, N. M., Garruss, A. S., Soenksen, L. R., Church, G. & Collins, J. J. A deep learning approach to programmable RNA switches. Nat. Commun. 11, 1–12 (2020).
Valeri, J. A. et al. Sequence-to-function deep learning frameworks for engineered riboregulators. Nat. Commun. 11, 1–14 (2020).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
Renaud, N. et al. Deeprank: A deep learning framework for data mining 3d protein–protein interfaces. Nat. Commun. 12, 1–8 (2021).
Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell 180, 688–702 (2020).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: Generative models for matter engineering. Science 361, 360–365 (2018).
Teva, et al. Global and China Drug Repositioning Market Size, Status and Forecast 2020–2027 (QYResearch Group, 0AD).
Rifaioglu, A. S. et al. Deepscreen: High performance drug-target interaction prediction with convolutional neural networks using 2-d structural compound representations. Chem. Sci. 11, 2531–2557 (2020).
Ruiz Puentes, P. et al. Pharmanet: Pharmaceutical discovery with deep recurrent neural networks. PLOS ONE 16, 1–22. https://doi.org/10.1371/journal.pone.0241728 (2021).
Scantlebury, J., Brown, N., Von Delft, F. & Deane, C. M. Data set augmentation allows deep learning-based virtual screening to better generalize to unseen target classes and highlight important binding interactions. J. Chem. Inf. Model. 60, 3722–3730 (2020).
Liao, Z. et al. Deepdock: Enhancing ligand–protein interaction prediction by a combination of ligand and structure information. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 311–317 (IEEE, 2019).
Torng, W. & Altman, R. B. Graph convolutional neural networks for predicting drug–target interactions. J. Chem. Inf. Model. 59, 4131–4149 (2019).
Lim, J. et al. Predicting drug–target interaction using a novel graph neural network with 3d structure-embedded graph representation. J. Chem. Inf. Model. 59, 3981–3988 (2019).
Zheng, S., Li, Y., Chen, S., Xu, J. & Yang, Y. Predicting drug–protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2, 134–140 (2020).
Li, G., Müller, M., Thabet, A. & Ghanem, B. Deepgcns: Can GCNS go as deep as CNNS? In The IEEE International Conference on Computer Vision (ICCV) (2019).
Li, G., Xiong, C., Thabet, A. & Ghanem, B. Deepergcn: All you need to train deeper GCNS. arXiv (2020). http://arxiv.org/abs/arXiv:2006.07739.
Duvenaud, D. K. et al. Convolutional networks on graphs for learning molecular fingerprints. Adv. Neural Inf. Process. Syst. 28 (2015).
Feinberg, E. N. et al. PotentialNet for molecular property prediction. ACS Central Sci. 4, 1520–1530. https://doi.org/10.1021/acscentsci.8b00507 (2018).
Feinberg, E. N., Joshi, E., Pande, V. S. & Cheng, A. C. Improvement in ADMET prediction with multitask deep featurization. J. Med. Chem. 63, 8835–8848. https://doi.org/10.1021/acs.jmedchem.9b02187 (2020).
Goodfellow, I. J., Shlens, J. & . Szegedy, C. Explaining and harnessing adversarial examples. In International Conference on Learning Representations (2015).
Engstrom, L. et al. Learning perceptually-aligned representations via adversarial robustness ArXiv preprint http://arxiv.org/abs/arXiv:1906.00945 (2019).
Mądry, A., Makelov, A., Schmidt, L., Tsipras, D. & Vladu, A. Towards deep learning models resistant to adversarial attacks. In International Conference on Learning Representations (2017).
Dai, H. et al. Adversarial attack on graph structured data. In International Conference on Machine Learning, 1115–1124 (PMLR, 2018).
Mendez, D. et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2019).
Corsello, S. M. et al. The drug repurposing hub: A next-generation drug library and information resource. Nat. Med. 23, 405–408 (2017).
Chen, L. et al. Hidden bias in the dud-e dataset leads to misleading performance of deep learning in structure-based virtual screening. PloS ONE 14 (2019).
Kuroda, M. A novel descriptor based on atom-pair properties. J. Cheminform. 9. https://doi.org/10.1186/s13321-016-0187-6 (2017).
Sheridan, R. P. et al. Experimental error, kurtosis, activity cliffs, and methodology: What limits the predictivity of quantitative structure–activity relationship models?. J. Chem. Inf. Model. 60, 1969–1982. https://doi.org/10.1021/acs.jcim.9b01067 (2020).
Byron Carpenter, G. L. Human adenosine a2a receptor: Molecular mechanism of ligand binding and activation. Front. Pharmacol. 8, 615–629 (2017).
Cristalli, G. et al. Adenosine deaminase inhibitors: Synthesis and structure–activity relationships of 2-hydroxy-3-nonyl derivatives of azoles. J. Med. Chem. 37, 201–205 (1994).
Taira, K. & Benkovic, S. J. Evaluation of the importance of hydrophobic interactions in drug binding to dihydrofolate reductase. J. Med. Chem. 31, 129–137 (1988).
Tsou, H.-R. et al. Optimization of 6, 7-disubstituted-4-(arylamino) quinoline-3-carbonitriles as orally active, irreversible inhibitors of human epidermal growth factor receptor-2 kinase activity. J. Med. Chem. 48, 1107–1131 (2005).
Luft, F. C. 11\(\beta\)-hydroxysteroid dehydrogenase-2 and salt-sensitive hypertension. Circulation 133, 1335–1337 (2016).
Steffan, R. J. et al. Synthesis and activity of substituted 4-(indazol-3-yl) phenols as pathway-selective estrogen receptor ligands useful in the treatment of rheumatoid arthritis. J. Med. Chem. 47, 6435–6438 (2004).
Neyshabur, B., Bhojanapalli, S., Mcallester, D. & Srebro, N. Exploring generalization in deep learning. In Advances in Neural Information Processing Systems, (eds. Guyon, I. et al.), Vol. 30 (Curran Associates, Inc., 2017).
Ohno, K., Mori, K., Orita, M. & Takeuchi, M. Computational insights into binding of bisphosphates to farnesyl pyrophosphate synthase. Curr. Med. Chem. 18, 220–233 (2011).
Malanovic, N. et al. S-adenosyl-l-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: Implications for homocysteine as a risk factor of atherosclerosis. J. Biol. Chem. 283 (2008).
Lucas, R. et al. Synthesis and enzyme inhibitory activities of a series of lipidic diamine and aminoalcohol derivatives on cytosolic and secretory phospholipases a2. Bioorgan. Med. Chem. Lett. 10, 285–288 (2000).
Kayhan, N. et al. The adenosine deaminase inhibitor erythro-9-[2-hydroxyl-3-nonyl]-adenine decreases intestinal permeability and protects against experimental sepsis: A prospective, randomised laboratory investigation. Critical Care 12, 1–11 (2008).
McKenna, R., Neidle, S. & Serafinowski, P. Structure of 5′-chloro-3′,5′-dideoxyformycin a monohydrate. the effects of protonation on formycin structure and conformation. Acta Crystallogr. 46, 2448–2450 (1990).
Lerner, L. M. & Rossi, R. R. Inhibition of adenosine deaminase by alcohols derived from adenine nucleosides. Biochemistry 11 (1972).
Mysinger, M. M., Carchia, M., Irwin, J. J. & Shoichet, B. K. Directory of useful decoys, enhanced (dud-e): Better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012).
Cleves, A. E. & Jain, A. N. Structure-and ligand-based virtual screening on dud-e+: Performance dependence on approximations to the binding pocket. J. Chem. Inf. Model. 60, 4296–4310 (2020).
Chaput, L., Martinez-Sanz, J., Saettel, N. & Mouawad, L. Benchmark of four popular virtual screening programs: Construction of the active/decoy dataset remains a major determinant of measured performance. J. Cheminform. 8, 1–17 (2016).
Yu, F. & Koltun, V. Multi-scale context aggregation by dilated convolutions. CoRR. http://arxiv.org/abs/arXiv:1511.07122 (2016).
Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996).
Sanders, J. M. et al. Pyridinium-1-yl bisphosphonates are potent inhibitors of farnesyl diphosphate synthase and bone resorption. J. Med. Chem. 48, 2957–2963. https://doi.org/10.1021/jm040209d (2005).
Sanders, J. M. et al. 3-d QSAR investigations of the inhibition of leishmania major farnesyl pyrophosphate synthase by bisphosphonates. J. Med. Chem. 46, 5171–5183. https://doi.org/10.1021/jm0302344 (2003).
Szajnman, S. H. et al. Synthesis and biological evaluation of 2-alkylaminoethyl-1, 1-bisphosphonic acids against trypanosoma cruzi and toxoplasma gondii targeting farnesyl diphosphate synthase. Bioorgan. Med. Chem. 16, 3283–3290. https://doi.org/10.1016/j.bmc.2007.12.010 (2008).
Simoni, D. et al. Design, synthesis, and biological evaluation of novel aminobisphosphonates possessing an in vivo antitumor activity through a y\(\delta\)-t lymphocytes-mediated activation mechanism. J. Med. Chem. 51, 6800–6807. https://doi.org/10.1021/jm801003y (2008).
Kotsikorou, E. & Oldfield, E. A quantitative structure-activity relationship and pharmacophore modeling investigation of aryl-x and heterocyclic bisphosphonates as bone resorption agents. J. Med. Chem. 46, 2932–2944. https://doi.org/10.1021/jm030054u (2003).
Castinetti, F., Conte-Devolx, B. & Brue, T. Medical treatment of cushing’s syndrome: Glucocorticoid receptor antagonists and mifepristone. Neuroendocrinology 92, 125–130. https://doi.org/10.1159/000314224 (2010).
Munro, D. D. & Wilson, L. Clobetasone butyrate, a new topical corticosteroid: Clinical activity and effects on pituitary-adrenal axis function and model of epidermal atrophy. BMJ 3, 626–628. https://doi.org/10.1136/bmj.3.5984.626 (1975).
Halobetasol propionate. Website (2021).
Wang, A. L. et al. Drug repurposing to treat glucocorticoid resistance in asthma. J. Person. Med. 11, 175. https://doi.org/10.3390/jpm11030175 (2021).
Process and intermediates for the synthesis of 8-[1-(3,5-bis-(trifluoromethyl)phenyl)-ethoxy-methyl]-8-phenyl-1,7-diaza-spiro[4.5]decan-2-one compounds. Website (2021).
Coghlan, M. J. et al. Synthesis and characterization of non-steroidal ligands for the glucocorticoid receptor: Selective quinoline derivatives with prednisolone-equivalent functional activity. J. Med. Chem. 44, 2879–2885. https://doi.org/10.1021/jm010228c (2001).
Clark, R. D. et al. 2-Benzenesulfonyl-8a-benzyl-hexahydro-2h-isoquinolin-6-ones as selective glucocorticoid receptor antagonists. Bioorgan. Med. Chem. Lett. 17, 5704–5708. https://doi.org/10.1016/j.bmcl.2007.07.055 (2007).
Clark, R. D. et al. 1h-Pyrazolo[3, 4-g]hexahydro-isoquinolines as selective glucocorticoid receptor antagonists with high functional activity. Bioorgan. Med. Chem. Lett. 18, 1312–1317. https://doi.org/10.1016/j.bmcl.2008.01.027 (2008).
Tegley, C. M. et al. 5-Benzylidene 1, 2-dihydrochromeno[3, 4-f]quinolines, a novel class of nonsteroidal human progesterone receptor agonists. J. Med. Chem. 41, 4354–4359. https://doi.org/10.1021/jm980366a (1998).
Kinney, W. A. et al. Bioisosteric replacement of the .alpha.-amino carboxylic acid functionality in 2-amino-5-phosphonopentanoic acid yields unique 3, 4-diamino-3-cyclobutene-1, 2-dione containing NMDA antagonists. J. Med. Chem. 35, 4720–4726. https://doi.org/10.1021/jm00103a010 (1992).
Huang, Y. H., Sinha, S. R., Fedoryak, O. D., Ellis-Davies, G. C. R. & Bergles, D. E. Synthesis and characterization of 4-methoxy-7-nitroindolinyl-d-aspartate, a caged compound for selective activation of glutamate transporters and N-methyl-d-aspartate receptors in brain tissue. Biochemistry 44, 3316–3326. https://doi.org/10.1021/bi048051m (2005).
Schoepp, D. D. et al. D, l-(tetrazol-5-yl) glycine: A novel and highly potent NMDA receptor agonist. Eur. J. Pharmacol. 203, 237–243. https://doi.org/10.1016/0014-2999(91)90719-7 (1991).
Dolman, N. P. et al. Synthesis and pharmacology of willardiine derivatives acting as antagonists of kainate receptors. J. Med. Chem. 48, 7867–7881. https://doi.org/10.1021/jm050584l (2005).
Chin, A. C., Yovanno, R. A., Wied, T. J., Gershman, A. & Lau, A. Y. D-serine potently drives ligand-binding domain closure in the ionotropic glutamate receptor GluD2. Structure 28, 1168-1178.e2. https://doi.org/10.1016/j.str.2020.07.005 (2020).
Butini, S. et al. 1h-Cyclopentapyrimidine-2, 4(1h, 3h)-dione-related ionotropic glutamate receptors ligands. Structure-activity relationships and identification of potent and selective iGluR5 modulators. J. Med. Chem. 51, 6614–6618. https://doi.org/10.1021/jm800865a (2008).
Ziemińska, E., Stafiej, A. & Łazarewicz, J. W. Role of group I metabotropic glutamate receptors and NMDA receptors in homocysteine-evoked acute neurodegeneration of cultured cerebellar granule neurones. Neurochem. Int. 43, 481–492. https://doi.org/10.1016/s0197-0186(03)00038-x (2003).
Kozikowski, A. P., Tuckmantel, W., Reynolds, I. J. & Wroblewski, J. T. Synthesis and bioactivity of a new class of rigid glutamate analogs. Modulators of the n-methyl-d-aspartate receptor. J. Med. Chem. 33, 1561–1571. https://doi.org/10.1021/jm00168a007 (1990).
Faria, M. et al. Liquid chromatography–tandem mass spectrometry method for quantification of thymidine kinase activity in human serum by monitoring the conversion of 3′-deoxy-3′-fluorothymidine to 3′-deoxy-3′-fluorothymidine monophosphate. J. Chromatogr. B 907, 13–20. https://doi.org/10.1016/j.jchromb.2012.08.024 (2012).
Bridges, E. G., Selden, J. R. & Luo, S. Nonclinical safety profile of telbivudine, a novel potent antiviral agent for treatment of hepatitis b. Antimicrobial Agents Chemother. 52, 2521–2528. https://doi.org/10.1128/aac.00029-08 (2008).
Kulikowski, T. Structure–activity relationships and conformational features of antiherpetic pyrimidine and purine nucleoside analogues. A review. Pharm. World Sci. 16, 127–138. https://doi.org/10.1007/bf01880663 (1994).
Chong, Y. & Chu, C. K. Understanding the unique mechanism of l-FMAU (clevudine) against hepatitis b virus: Molecular dynamics studies. Bioorgan. Med. Chem. Lett. 12, 3459–3462. https://doi.org/10.1016/s0960-894x(02)00747-3 (2002).
Suzuki, N. Mode of action of trifluorothymidine (TFT) against DNA replication and repair enzymes. Int. J. Oncol.https://doi.org/10.3892/ijo.2011.1003 (2011).
Manikowski, A. et al. Inhibition of herpes simplex virus thymidine kinases by 2-phenylamino-6-oxopurines and related compounds: Structure–activity relationships and antiherpetic activity in vivo. J. Med. Chem. 48, 3919–3929. https://doi.org/10.1021/jm049059x (2005).
Xu, H. et al. Synthesis, properties, and pharmacokinetic studies of n2-phenylguanine derivatives as inhibitors of herpes simplex virus thymidine kinases. J. Med. Chem. 38, 49–57. https://doi.org/10.1021/jm00001a010 (1995).
Balzarini, J., Bohman, C. & Clercq, E. D. Differential mechanism of cytostatic effect of (e)-5-(2-bromovinyl)-2-deoxyuridine, 9-(1, 3-dihydroxy-2-propoxymethyl)guanine, and other antiherpetic drugs on tumor cells transfected by the thymidine kinase gene of herpes simplex virus type 1 or type 2. J. Biol. Chem. 268, 6332–6337. https://doi.org/10.1016/s0021-9258(18)53257-9 (1993).
Krenitsky, T. A., Elion, G. B., Henderson, A. M. & Hitchings, G. H. Inhibition of human purine nucleoside phosphorylase. J. Biol. Chem. 243, 2876–2881. https://doi.org/10.1016/s0021-9258(18)93353-3 (1968).
Koellner, G., Luić, M., Shugar, D., Saenger, W. & Bzowska, A. Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 resolution. J. Mol. Biol. 265, 202–216. https://doi.org/10.1006/jmbi.1996.0730 (1997).
Lee, S. H. & Sartorelli, A. C. Conversion of 6-thioguanine to the nucleoside level by purine nucleoside phosphorylase of sarcoma 180 and sarcoma 180/tg ascites cells. Cancer Res. 41, 1086–1090 (1981).
An enzymatic synthesis of nucleosides of n2-acetyl-o6-[2-(4-nitrophenyl)ethyl]guanine. Website (2021).
Bzowska, A., Kulikowska, E. & Shugar, D. Linear free energy relationships for n(7)-substituted guanosines as substrates of calf spleen purine nucleoside phosphorylase. possible role of n(7)-protonation as an intermediary in phosphorolysis. Z. Naturforschung C 48, 803–811. https://doi.org/10.1515/znc-1993-9-1020 (1993).
Chaban, T. et al. Thiazolo[5,4-d]pyrimidines and thiazolo[4,5-d] pyrimidines: Review on synthesis and pharmacological importance of their derivatives. Pharmacia 65, 54–70 (2018).
Koellner, G., Stroh, A., Raszewski, G., Holý, A. & Bzowska, A. Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with multisubstrate analogue inhibitor with 2, 6-diaminopurine aglycone. Nucleosides Nucleotides Nucleic Acids 22, 1699–1702. https://doi.org/10.1081/ncn-120023117 (2003).
López-Lira, C. et al. New benzimidazolequinones as trypanosomicidal agents. Bioorgan. Chem. https://doi.org/10.1016/j.bioorg.2021.104823 (2021).
Stoeckler, J. D., Cambor, C., Kuhns, V., Shih-Hsi, C. & Parks, R. E. Inhibitors of purine nucleoside phosphorylase. Biochem. Pharmacol. 31, 163–171. https://doi.org/10.1016/0006-2952(82)90206-4 (1982).
Aury-Landas, J. et al. Anti-inflammatory and chondroprotective effects of the s-adenosylhomocysteine hydrolase inhibitor 3-deazaneplanocin a, in human articular chondrocytes. Sci. Rep. https://doi.org/10.1038/s41598-017-06913-6 (2017).
Yuan, K. et al. Comparative transcriptomics analysis of streptococcus mutans with disruption of LuxS/AI-2 quorum sensing and recovery of methyl cycle. Arch. Oral Biol. https://doi.org/10.1016/j.archoralbio.2021.105137 (2021).
Malladi, V. L., Sobczak, A. J., Meyer, T. M., Pei, D. & Wnuk, S. F. Inhibition of LuxS by s-ribosylhomocysteine analogues containing a [4-aza]ribose ring. Bioorgan. Med. Chem. 19, 5507–5519. https://doi.org/10.1016/j.bmc.2011.07.043 (2011).
Ueland, P. M. & Saebo, J. S-adenosylhomocysteinase from mouse liver. Effect of adenine and adenine nucleotides on the enzyme catalysis. Biochemistry 18, 4130–4135. https://doi.org/10.1021/bi00586a012 (1979).
Borcherding, D. R. et al. Potential inhibitors of s-adenosylmethionine-dependent methyltransferases. 11. Molecular dissections of neplanocin a as potential inhibitors of s-adenosylhomocysteine hydrolase. J. Med. Chem. 31, 1729–1738. https://doi.org/10.1021/jm00117a011 (1988).
Wolfe, M. S., Lee, Y., Bartlett, W. J., Borcherding, D. R. & Borchardt, R. T. 4-Modified analogs of aristeromycin and neplanocin a: Synthesis and inhibitory activity toward s-adenosyl-l-homocysteine hydrolase. J. Med. Chem. 35, 1782–1791. https://doi.org/10.1021/jm00088a013 (1992).
Liu, S., Sheng, Y. C. & Borchardt, R. T. Aristeromycin-5‘-carboxaldehyde: A potent inhibitor of s-adenosyl- l-homocysteine hydrolase. J. Med. Chem. 39, 2347–2353. https://doi.org/10.1021/jm950916u (1996).
Wnuk, S. F. et al. Nucleic acid-related compounds. 84. Synthesis of 6-(e and z)-halohomovinyl derivatives of adenosine, inactivation of s-adenosyl-l-homocysteine hydrolase, and correlation of anticancer and antiviral potencies with enzyme inhibition. J. Med. Chem. 37, 3579–3587. https://doi.org/10.1021/jm00047a015 (1994).
Ando, T. et al. Synthesis of 4′-modified noraristeromycins to clarify the effect of the 4′-hydroxyl groups for inhibitory activity against s-adenosyl-l-homocysteine hydrolase. Bioorgan. Med. Chem. Lett. 18, 2615–2618. https://doi.org/10.1016/j.bmcl.2008.03.029 (2008).
Mouchlis, V. D., Armando, A. & Dennis, E. A. Substrate-specific inhibition constants for phospholipase a2 acting on unique phospholipid substrates in mixed micelles and membranes using lipidomics. J. Med. Chem. 62, 1999–2007. https://doi.org/10.1021/acs.jmedchem.8b01568 (2019).
Dillard, R. D. et al. Indole inhibitors of human nonpancreatic secretory phospholipase a2. 2. Indole-3-acetamides with additional functionality. J. Med. Chem. 39, 5137–5158. https://doi.org/10.1021/jm960486n (1996).
Aid 720700 - fluorescence-based biochemical high throughput primary assay to identify inhibitors of phospholipase c isozymes (plc-gamma1). - pubchem. National Center for Biotechnology Information (2021).
Smart, B. P., Oslund, R. C., Walsh, L. A. & Gelb, M. H. The first potent inhibitor of mammalian group x secreted phospholipase a2: Elucidation of sites for enhanced binding. J. Med. Chem. 49, 2858–2860. https://doi.org/10.1021/jm060136t (2006).
Adlere, I. et al. Modulators of CXCR4 and CXCR7/ACKR3 function. Mol. Pharmacol. 96, 737–752. https://doi.org/10.1124/mol.119.117663 (2019).
Rosenberg, E. M. et al. Characterization, dynamics, and mechanism of CXCR4 antagonists on a constitutively active mutant. Cell Chem. Biol. 26, 662–673.e7. https://doi.org/10.1016/j.chembiol.2019.01.012 (2019).
Johnson, V. A. et al. Antiretroviral activity of AMD11070 (an orally administered CXCR4 entry inhibitor): Results of NIH/NIAID AIDS clinical trials group protocol a5210. AIDS Res. Hum. Retroviruses 35, 691–697. https://doi.org/10.1089/aid.2018.0256 (2019).
Jørgensen, A. S. et al. Biased action of the CXCR4-targeting drug plerixafor is essential for its superior hematopoietic stem cell mobilization. Commun. Biol.. https://doi.org/10.1038/s42003-021-02070-9 (2021).
Wilkinson, R. A. et al. Improved guanide compounds which bind the CXCR4 co-receptor and inhibit HIV-1 infection. Bioorgan. Med. Chem. Lett. 23, 2197–2201. https://doi.org/10.1016/j.bmcl.2013.01.107 (2013).
Thoma, G. et al. Orally bioavailable isothioureas block function of the chemokine receptor CXCR4 in vitro and in vivo. J. Med. Chem. 51, 7915–7920. https://doi.org/10.1021/jm801065q (2008).
Skerlj, R. et al. Synthesis and SAR of novel CXCR4 antagonists that are potent inhibitors of t tropic (x4) HIV-1 replication. Bioorgan. Med. Chem. Lett. 21, 262–266. https://doi.org/10.1016/j.bmcl.2010.11.023 (2011).
Chakraborty, S., Shah, N. H., Fishbein, J. C. & Hosmane, R. S. Investigations into specificity of azepinomycin for inhibition of guanase: Discrimination between the natural heterocyclic inhibitor and its synthetic nucleoside analogues. Bioorgan. Med. Chem. Lett. 22, 7214–7218. https://doi.org/10.1016/j.bmcl.2012.09.053 (2012).
Kasibhatla, S. R., Bookser, B. C., Probst, G., Appleman, J. R. & Erion, M. D. AMP deaminase inhibitors. 3. SAR of 3-(carboxyarylalkyl)coformycin aglycon analogues. J. Med. Chem. 43, 1508–1518. https://doi.org/10.1021/jm990448e (2000).
Bookser, B. C., Kasibhatla, S. R., Appleman, J. R. & Erion, M. D. AMP deaminase inhibitors. 2. Initial discovery of a non-nucleotide transition-state inhibitor series. J. Med. Chem. 43, 1495–1507. https://doi.org/10.1021/jm990447m (2000).
Acknowledgements
This work was partially supported by a Microsoft AI for Health computational grant. The authors would also like to thank Juan Camilo Pérez for his insights on adversarial data adaptation. Natalia Valderrama and Isabela Hernández acknowledge the support of UniAndes-DeepMind Scholarship 2021. The authors would like to thank the Vice Presidency for Research & Creation’s Publication Fund at Universidad de los Andes for its financial support.
Author information
Authors and Affiliations
Contributions
P.A, C.G. conceptualization of the project. P.R., L.R.-G., N.V. and I.H. performed all the experiments, wrote the main manuscript and prepared all figures and tables. C.G. and L.D. supervised the experiments and review the manuscript. C.M.-C., J.C.C., P.A. supervised the project and reviewed the manuscript. All authors have revised and accepted the final version of the manuscript.
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruiz Puentes, P., Rueda-Gensini, L., Valderrama, N. et al. Predicting target–ligand interactions with graph convolutional networks for interpretable pharmaceutical discovery. Sci Rep 12, 8434 (2022). https://doi.org/10.1038/s41598-022-12180-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-12180-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.