Abstract
Phthiocerol dimycocerosate (PDIM) is an essential virulence lipid of Mycobacterium tuberculosis. In vitro culturing rapidly selects for spontaneous PDIM-negative mutants that have attenuated virulence and increased cell wall permeability, thus impacting the relevance of experimental findings. PDIM loss can also reduce the efficacy of the BCG Pasteur vaccine. Here we show that vancomycin susceptibility can rapidly screen for M. tuberculosis PDIM production. We find that metabolic deficiency of methylmalonyl-CoA impedes the growth of PDIM-producing bacilli, selecting for PDIM-negative variants. Supplementation with odd-chain fatty acids, cholesterol or vitamin B12 restores PDIM-positive bacterial growth. Specifically, we show that propionate supplementation enhances PDIM-producing bacterial growth and selects against PDIM-negative mutants, analogous to in vivo conditions. Our study provides a simple approach to screen for and maintain PDIM production, and reveals how discrepancies between the host and in vitro nutrient environments can attenuate bacterial pathogenicity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
WGS data have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA923717. A complete list of strains sequenced in this study and SRA accession numbers are given in Supplementary Table 11. Raw metabolomics data are provided in Supplementary Data 1. Source data are provided with this paper.
Code availability
WGS data were processed on the Albert Einstein College of Medicine High-Performance Computing Core (HPC). Scripts used for reference-guided assembly and variant calling are available via https://github.com/cvmulholland/MtbShortReadWGS.
References
Daffe, M. & Laneelle, M. A. Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 134, 2049–2055 (1988).
Rens, C., Chao, J. D., Sexton, D. L., Tocheva, E. I. & Av-Gay, Y. Roles for phthiocerol dimycocerosate lipids in Mycobacterium tuberculosis pathogenesis. Microbiology https://doi.org/10.1099/mic.0.001042 (2021).
Domenech, P. & Reed, M. B. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology 155, 3532–3543 (2009).
Manjunatha, U. H. et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 431–436 (2006).
Kirksey, M. A. et al. Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity. Infect. Immun. 79, 2829–2838 (2011).
Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R. Jr. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 (1999).
Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B. & Guilhot, C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34, 257–267 (1999).
Goren, M. B., Brokl, O. & Schaefer, W. B. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: phthiocerol dimycocerosate and the attenuation indicator lipid. Infect. Immun. 9, 150–158 (1974).
Murry, J. P., Pandey, A. K., Sassetti, C. M. & Rubin, E. J. Phthiocerol dimycocerosate transport is required for resisting interferon-γ-independent immunity. J. Infect. Dis. 200, 774–782 (2009).
Day, T. A. et al. Mycobacterium tuberculosis strains lacking surface lipid phthiocerol dimycocerosate are susceptible to killing by an early innate host response. Infect. Immun. 82, 5214–5222 (2014).
Rousseau, C. et al. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell. Microbiol. 6, 277–287 (2004).
Wang, Q. et al. PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367, 1147–1151 (2020).
Camacho, L. R. et al. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276, 19845–19854 (2001).
Tran, V., Ahn, S. K., Ng, M., Li, M. & Liu, J. Loss of lipid virulence factors reduces the efficacy of the BCG vaccine. Sci. Rep. 6, 29076 (2016).
Soetaert, K. et al. Increased vancomycin susceptibility in mycobacteria: a new approach to identify synergistic activity against multidrug-resistant mycobacteria. Antimicrob. Agents Chemother. 59, 5057–5060 (2015).
Rodrigues, L., Viveiros, M. & Ainsa, J. A. Measuring efflux and permeability in mycobacteria. Methods Mol. Biol. 1285, 227–239 (2015).
Jain, M. et al. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc. Natl Acad. Sci. USA 104, 5133–5138 (2007).
Pandey, A. K. & Sassetti, C. M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl Acad. Sci. USA 105, 4376–4380 (2008).
Griffin, J. E. et al. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227 (2012).
Gopinath, K., Moosa, A., Mizrahi, V. & Warner, D. F. Vitamin B12 metabolism in Mycobacterium tuberculosis. Future Microbiol. 8, 1405–1418 (2013).
Gopinath, K. et al. A vitamin B12 transporter in Mycobacterium tuberculosis. Open Biol. 3, 120175 (2013).
Savvi, S. et al. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 190, 3886–3895 (2008).
Yang, X., Nesbitt, N. M., Dubnau, E., Smith, I. & Sampson, N. S. Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 48, 3819–3821 (2009).
Koh, E. I. et al. Chemical–genetic interaction mapping links carbon metabolism and cell wall structure to tuberculosis drug efficacy. Proc. Natl Acad. Sci. USA 119, e2201632119 (2022).
Quinonez, C. G. et al. The role of fatty acid metabolism in drug tolerance of Mycobacterium tuberculosis. mBio 13, e0355921 (2022).
Hicks, N. D. et al. Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3, 1032–1042 (2018).
Wang, H. et al. An essential bifunctional enzyme in Mycobacterium tuberculosis for itaconate dissimilation and leucine catabolism. Proc. Natl Acad. Sci. USA 116, 15907–15913 (2019).
Ortalo-Magne, A. et al. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 178, 456–461 (1996).
Jain, P. et al. Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio 5, e01245–01214 (2014).
Dechow, S. J., Baker, J. J., Murto, M. & Abramovitch, R. B. ppe51 variants enable growth of Mycobacterium tuberculosis at acidic pH by selectively promoting glycerol uptake. J. Bacteriol. https://doi.org/10.1128/jb.00212-22 (2022).
Gopal, P. et al. Pyrazinamide resistance is caused by two distinct mechanisms: prevention of coenzyme A depletion and loss of virulence factor synthesis. ACS Infect. Dis. 2, 616–626 (2016).
Orsi, R. H., Bowen, B. M. & Wiedmann, M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics 11, 102 (2010).
Mizrahi, V. & Andersen, S. J. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol. Microbiol. 29, 1331–1339 (1998).
Dolan, S. K. et al. Loving the poison: the methylcitrate cycle and bacterial pathogenesis. Microbiology 164, 251–259 (2018).
Munoz-Elias, E. J., Upton, A. M., Cherian, J. & McKinney, J. D. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122 (2006).
Lee, W., VanderVen, B. C., Fahey, R. J. & Russell, D. G. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 288, 6788–6800 (2013).
Singh, A. et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5, e1000545 (2009).
Lu, R. et al. Catabolism of the cholesterol side chain in Mycobacterium tuberculosis is controlled by a redox-sensitive thiol switch. ACS Infect. Dis. 3, 666–675 (2017).
Eoh, H. & Rhee, K. Y. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc. Natl Acad. Sci. USA 111, 4976–4981 (2014).
Dulberger, C. L., Rubin, E. J. & Boutte, C. C. The mycobacterial cell envelope—a moving target. Nat. Rev. Microbiol. 18, 47–59 (2020).
Marrero, J., Rhee, K. Y., Schnappinger, D., Pethe, K. & Ehrt, S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl Acad. Sci. USA 107, 9819–9824 (2010).
Block, A. M. et al. Mycobacterium tuberculosis requires the outer membrane lipid phthiocerol dimycocerosate for starvation-induced antibiotic tolerance. mSystems 8, e0069922 (2023).
Maksymiuk, C. et al. Comparison of transposon and deletion mutants in Mycobacterium tuberculosis: the case of rv1248c, encoding 2-hydroxy-3-oxoadipate synthase. Tuberculosis 95, 689–694 (2015).
Chen, J. M., Islam, S. T., Ren, H. & Liu, J. Differential productions of lipid virulence factors among BCG vaccine strains and implications on BCG safety. Vaccine 25, 8114–8122 (2007).
Bloch, H. & Segal, W. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72, 132–141 (1956).
Babunovic, G. H. et al. CRISPR interference reveals that all-trans-retinoic acid promotes macrophage control of Mycobacterium tuberculosis by limiting bacterial access to cholesterol and propionyl coenzyme A. mBio 13, e0368321 (2022).
Dubos, R. J. & Middlebrook, G. Media for tubercle bacilli. Am. Rev. Tuberc. 56, 334–345 (1947).
Dubos, R. J. Rapid and submerged growth of mycobacteria in liquid media. Proc. Soc. Exp. Biol. Med. 58, 361–362 (1945).
Li, S. et al. CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nat. Microbiol. 7, 766–779 (2022).
Xu, W. et al. Chemical genetic interaction profiling reveals determinants of intrinsic antibiotic resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.01334-17 (2017).
Chengalroyen, M. D. et al. DNA-dependent binding of nargenicin to DnaE1 inhibits replication in Mycobacterium tuberculosis. ACS Infect. Dis. 8, 612–625 (2022).
Wang, Q. & Boshoff, H. I. M. Determining minimum inhibitory concentrations in liquid cultures or on solid medium. Methods Mol. Biol. 2314, 595–609 (2021).
Chandra, P., Grigsby, S. J. & Philips, J. A. Immune evasion and provocation by Mycobacterium tuberculosis. Nat. Rev. Microbiol. 20, 750–766 (2022).
DeJesus, M. A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio https://doi.org/10.1128/mBio.02133-16 (2017).
Bosch, B. et al. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell 184, 4579–4592 e4524 (2021).
Zhang, Y. J. et al. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155, 1296–1308 (2013).
Sambandamurthy, V. K. et al. Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent andimmunocompromised mice against experimental tuberculosis. Vaccine 24, 6309–6320 (2006).
Jain, P. et al. φ(2)GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J. Clin. Microbiol. 50, 1362–1369 (2012).
Vilcheze, C. et al. Rational design of biosafety level 2-approved, multidrug-resistant strains of Mycobacterium tuberculosis through nutrient auxotrophy. mBio https://doi.org/10.1128/mBio.00938-18 (2018).
Jacobs, W. R., Jr. & Tiwari, S. Double auxotrophic and uses thereof. US patent 11666648 (2023).
Stover, C. K. et al. New use of BCG for recombinant vaccines. Nature 351, 456–460 (1991).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Dai, Y. & Hsiao, J. J. Discovery Metabolomics LC/MS Methods Optimized for Polar Metabolites Application Note (Agilent Technologies, 2019).
National Research Council of the National Academies Guide for the Care and Use of Laboratory Animals 8th edn. (National Academies Press, 2011).
Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 56, 2.4.1–2.4.5 (2001).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience https://doi.org/10.1093/gigascience/giab008 (2021).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Okonechnikov, K., Conesa, A. & Garcia-Alcalde, F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Acknowledgements
We thank B. Chen, J. Kim and M. Chen for assistance with animal experiments; A. Zhi Dai for technical support; S. Tiwari who constructed the mc28398 mutant; and the laboratories of J. Rock, Rockefeller University, NY, and J. Chan, Rutgers University, NJ, for their feedback and independent validation of VAN-P PDIM assays. We acknowledge support from the following grants: National Institutes of Health/National Institute of Allergy and Infectious Diseases R01 AI139465 for C.V.M., T.J.W., J.C., E.Z.R.-F. and M.B.; R01 AI175972 for C.V.M., T.J.W., J.C., E.Z.R.-F. and M.B.; and AI026170 for C.V., S.R. and W.R.J.; the Potts Memorial Foundation for C.V.M., E.Z.R.-F. and M.B.; Albert Einstein College of Medicine internal funding for E.Z.R.-F. and M.B.; the Institutional AIDS training grant, Training in HIV/AIDS Pathogenesis; Basic and Translational Research (T32 AI007501) for M.W.S.; and the Albert Einstein College of Medicine MSTP training grant T32GM149364 for M.W.S.
Author information
Authors and Affiliations
Contributions
C.V.M. and M.B. conceived and designed the study. C.V.M., T.J.W., J.C., C.V., S.R., M.W.S. and E.Z.R.-F. performed the experiments. C.V.M., T.J.W., C.V., M.W.S. and M.B. analysed the data. M.B. and W.R.J. provided resources. C.V.M. and M.B. wrote the paper. T.J.W., C.V., S.R., M.W.S., E.Z.R.-F. and W.R.J. critically reviewed and edited the paper.
Corresponding author
Ethics declarations
Competing interests
C.V.M. and M.B. are inventors on a pending patent related to this work (US Patent Application Number 63/527,831, filed 20 July 2023). They declare that they have no other competing interests. The other authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Hyungjin Eoh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Resistance of PDIM(-) and PDIM(+) Mtb to high molecular weight compounds.
a–g, MIC assays of Mtb mc27902 [PDIM(+)] and mc28398 [PDIM(-)] to (a) ramoplanin (RAM), (b) teicoplanin (TEC), (c) vancomycin (VAN), (d) rifampicin (RIF), (e) azithromycin (AZM), (f) erythromycin (ERY), and (g) isoniazid (INH). Compounds are arranged by descending molecular weight, which is shown on the MIC plots. MICs were performed in 7H9/OADC/glycerol/tyloxapol + PALM media and bacterial growth was measured after 10 days of incubation and normalized to drug-free controls. Mean ± s.d. for n = 4 biological replicates from two independent experiments. h, Ethidium Bromide uptake of mc27902 and mc28398. Uptake in whole cell suspensions was monitored by fluorescence (Ex 355 nm/Em 590 nm). Mean ± s.d. for n = 4 biological replicates, each measured in five technical replicates. Uptake data are representative of two independent experiments. *P < 0.001; two-way ANOVA with Šidák’s multiple comparison test.
Extended Data Fig. 2 Propionate and vitamin B12 supplementation selectively increase vancomycin resistance of PDIM(+) Mtb improving assay robustness and reducing time to result.
a, Vancomycin MICs for the PDIM reference strain set in standard 7H9/OADC/glycerol/tyloxapol + PALM media and additionally supplemented with 0.1 mM propionate. Growth was measured after 7, 10, and 14 days as indicated. Mean ± s.d. for n = 4 biological replicates from two independent experiments. b, Vancomycin MICs in standard media and additionally supplemented with 0.1 or 1.0 mM propionate or 7.4 μM vitamin B12. Growth was measured after 10 days. Mean ± s.d. for n = 4 biological replicates from two independent experiments. c, VAN10 assays in standard and supplemented media. Growth was measured after 10 days. Mean ± s.d. for n = 3 independent experiments, each performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; two-way ANOVA with Šidák’s multiple comparison test. The day seven data in (a) are also shown in Fig. 1c and are shown here alongside additional time points. The data in (b) includes one of the same experiments shown in (a), together with data from an independent experiment. The VAN10-P (+0.1 mM propionate) data in (c) are also shown in Fig. 1e and are shown here alongside additional conditions.
Extended Data Fig. 3 Tween 80 decreases vancomycin resistance and abolishes PDIM-related differences in MIC.
a, VAN-P MICs for PDIM(+) and PDIM(-) Mtb H37Rv using either tyloxapol or Tween 80 as the culture detergent. b, Vancomycin MICs for PDIM(+) H37Rv wildtype in standard 7H9/OADC/glycerol media and supplemented with propionate or vitamin B12 using either tyloxapol or Tween 80 as the detergent. Mean ± s.d. for n = 4 biological replicates from two independent experiments.
Extended Data Fig. 4 VAN-P assays predict PDIM levels across different Mtb strains and lineages and enable re-isolation of single PDIM(+) clones.
a, VAN10-P assays for a range of virulent Mtb strains belonging to different lineages. Bacterial growth was measured after 7, 10, and 14 days of incubation as indicated. Data are from the same experiment in Fig. 2a and show additional time points plus an independent experimental repeat measured on day 10 (hatched bars with unfilled symbols). b, VAN10-P assays for strains with an H37Rv background including mc27902 and mc28398. H37Rv-SC is a single PDIM(+) clone isolated from H37Rv-B by VAN10-P colony screening. This clone was used as our PDIM(+) H37Rv wildtype strain throughout this work and was used to construct H37Rv ΔppsD and ΔppsD::comp mutants (Supplementary Table 2). Data in (a,b) show mean ± s.d. for n = 9 pairwise comparisons between triplicate wells, except for the day 10 repeat in (a) where n = 4 pairwise comparisons between duplicate wells. c, VAN-P MICs of H37Rv stocks and H37Rv-SC. d, VAN-P MICs of non-H37Rv strains from (a). e–g, VAN-P MICs of single PDIM(+) clones isolated from Rag-/- mice using VAN10-P colony screening for (e) Erdman, (f) HN878, and (g) CDC1551 (see also Supplementary Table 3). Data are plotted together with MIC data from (d) for comparison. MIC data in (c–g) show mean ± s.d. for n = 4 biological replicates from two independent experiments. h–j, Determination that our Mtb mc26230 stock is a mixed population and re-isolation of a single PDIM(+) clone by VAN10-P screening. (h) VAN10-P assay of single colonies isolated from our mc26230 stock (n = 40) and (i) following a single passage in 10 μg/ml vancomycin (n = 20). Vancomycin significantly enriched for PDIM(+) bacilli (P < 0.0001 two-tailed Mann-Whitney test), facilitating re-isolation of low-frequency PDIM(+) clones. Each colony was assayed in triplicate and data points represent mean VAN10-P growth%. Lines indicate the median. j, VAN-P MICs of PDIM(+) (AE1601) and PDIM(-) (AE1611) mc26230 clones identified by VAN10-P colony screening (see also Supplementary Table 3). Mean ± s.d. for n = 6 biological replicates from two independent experiments.
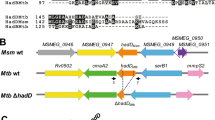
Extended Data Fig. 5 Assessment of ppsC homopolymeric tract mutations.
a, Schematic showing the location of a homopolymeric tract region in the ppsC gene. Sequence inserts show two adjacent 7-cytosine homopolymeric tracts (c.2668 and c.2685) ± 5 bp on either side. Numbers in black indicate the position in the ppsC gene and numbers in red the genomic position in the H37Rv genome. b–d, Analysis of the ppsC homopolymeric tract region in Δtgs1 mutants and identification of frameshift mutations. WGS variant calling failed to identify PDIM mutations in Δtgs1-5, Δtgs1-8 and Δtgs1-9 despite a PDIM(-) result in VAN-P MICs (Fig. 2b) and validation of Δtgs1-9 as PDIM(-) by TLC (Fig. 2c). Close manual inspection of WGS reads showed the ppsC homopolymeric tract region is poorly covered by Illumina MiSeq and identified potentially missed variant calls. PCR and Sanger sequencing confirmed the presence of a 2668(C)7→6 frameshift mutation in both Δtgs1-5 (b) and Δtgs1-9 (d) and identified a 2668(C)7→8 mutation in Δtgs1-8 that was not covered at all by WGS (c). (b–d) were created with Geneious Prime® 2022.2.2 and Illustrator 26.4.1. Coverage has been cropped to a read depth of 60 ×. See also Supplementary Table 5.
Extended Data Fig. 6 Effect of different media supplements on growth of PDIM(+) and PDIM(-) Mtb.
a, Growth of PDIM(+) and PDIM(-) Mtb H37Rv in standard 7H9/OADC/glycerol/tyloxapol and b–j, the same media with additional supplements as indicated. k–m, Growth using Tween 80 instead of tyloxapol as the culture detergent with additional supplements as indicated. Mean ± s.d. for n = 3 biological replicates. Data are representative of at least two independent experiments. (a, b, d, f) show independent experimental repeats for the conditions in Fig. 3b,c. *P < 0.001 for both wt and comp versus ΔppsD; two-way ANOVA with Tukey’s multiple comparison test. For some data points the s.d. is smaller than the data symbols.
Extended Data Fig. 7 Effects of propionate and vitamin B12 supplementation on MMCoA and propionyl-CoA metabolic pathways in Mtb.
a, Abundance of metabolites in propionyl-CoA and MMCoA metabolism in PDIM(+) Mtb H37Rv wildtype grown in standard 7H9/OADC/glycerol/tyloxapol media and supplemented with propionate or vitamin B12, and b, in PDIM(+) and PDIM(-) H37Rv grown in 7H9/OADC/glycerol/tyloxapol ± 0.1 mM propionate. Abundances are shown as normalized area under the curve (AUC) (see Methods). Mean ± s.d. for n = 6 biological replicates from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; one-way ANOVA with Tukey’s multiple comparison test. Significant differences compared to unsupplemented media are indicated in (a), and between ± propionate for each strain and between strains for each condition in (b). PROP, propionate; PROP-CoA, propionyl-CoA; MMCoA, methylmalonyl-CoA; SUC-CoA, succinyl-CoA; SUC, succinate; 2MC/2MIC, 2-methyl(iso)citrate; and PYR, pyruvate. Succinyl-CoA and methyl(iso)citrate were not able to be detected in samples by our method. Propionyl-CoA was close to the detection limit and was not detected in all samples (n.d. = not detected). The data for MMCoA are also shown in Fig. 3d,e. See also Supplementary Fig. 8.
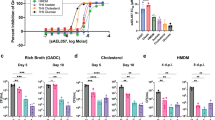
Extended Data Fig. 8 Propionate and vitamin B12 supplementation prevent PDIM loss in Mtb.
a, Schematic overview of in vitro evolution experiments. Triplicate inkwells containing standard 7H9/OADC/glycerol/tyloxapol or media supplemented with propionate or vitamin B12 were inoculated with frozen Mtb culture stock (P0) and incubated for 7-10 days (P1). Cultures were then diluted into fresh media every 7 days for serial passage (P2 to PX). Selected passages were input into VAN10-P assays at the time of passage to assess PDIM production over the course of the experiment. For TLC lipid analysis, frozen stocks were first outgrown in media without propionate or vitamin B12 for a single passage to allow the strains to recover before 14C-labelling. Figure created with BioRender.com. b, TLC lipid analysis of H37Rv-B before and after six serial passages in ± 0.1 or 1.0 mM propionate. This figure shows the full TLC plate from Fig. 4a with results for both biological replicates analysed by TLC. c, VAN10-P assays for H37Rv-SC [PDIM(+) H37Rv wildtype] passaged in ± 0.1 mM propionate. d, H37Rv-A and e, H37Rv-B passaged in ± 7.4 μM vitamin B12. Mean ± s.d. for n = 3 biological replicates, each assayed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; two-way ANOVA with Šidák’s (c) or Tukey’s (d, e) multiple comparison test. Significant differences between conditions are indicated in (c) and between timepoints in (d, e).
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1–11 and References.
Supplementary Data 1
Raw LC–MS metabolomics data.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed TLC image.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed TLC image.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed TLC image.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed TLC image.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mulholland, C.V., Wiggins, T.J., Cui, J. et al. Propionate prevents loss of the PDIM virulence lipid in Mycobacterium tuberculosis. Nat Microbiol (2024). https://doi.org/10.1038/s41564-024-01697-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41564-024-01697-8