Abstract
Different host plants represent ecologically dissimilar environments for phytophagous insects. The resulting divergent selection can promote the evolution of specialized host races, provided that gene flow is reduced between populations feeding on different plants. In black bean aphids belonging to the Aphis fabae complex, several morphologically cryptic taxa have been described based on their distinct host plant preferences. However, host choice and mate choice are largely decoupled in these insects: they are host-alternating and migrate between specific summer host plants and shared winter hosts, with mating occurring on the shared hosts. This provides a yearly opportunity for gene flow among aphids using different summer hosts, and raises the question if and to what extent the ecologically defined taxa are reproductively isolated. Here, we analyzed a geographically and temporally structured dataset of microsatellite genotypes from A. fabae that were mostly collected from their main winter host Euonymus europaeus, and additionally from another winter host and fourteen summer hosts. The data reveals multiple, strongly differentiated genetic clusters, which differ in their association with different summer and winter hosts. The clusters also differ in the frequency of infection with two heritable, facultative endosymbionts, separately hinting at reproductive isolation and divergent ecological selection. Furthermore, we found evidence for occasional hybridization among genetic clusters, with putative hybrids collected more frequently in spring than in autumn. This suggests that similar to host races in other phytophagous insects, both prezygotic and postzygotic barriers including selection against hybrids maintain genetic differentiation among A. fabae taxa, despite a common mating habitat.
Similar content being viewed by others
Introduction
Contrasting environments can impose differential selection on separate populations of a species, thereby causing ecologically based adaptive divergence. In this process of specialization, reduced gene flow and assortative mating may represent both drivers and effects of increasing population differentiation, and could eventually lay the ground for ecological speciation (e.g. Dobzhansky 1940; Rice 1987; Rundle and Nosil 2005; Schluter 2001). The evolution of separate, specialized species thereby represents the endpoint of a wide continuum of divergence, ranging from weak genetic differentiation to complete reproductive isolation between populations (Dobzhansky 1940; Nosil 2012; Schluter 2000). Among model organisms studied to investigate ecologically based population divergence and the potential of ecological speciation, phytophagous insects assume a prominent position (e.g. Berlocher and Feder 2002; Funk et al. 2002; Matsubayashi et al. 2010; Via 2001). Their host plants often represent habitat, food source, and mating site all in one, and the variable chemical and physical properties of different plant species may impose very specific selection pressures on the insects exploiting them. Examples of polyphagous insect species that appear structured into host-specialized lineages, often referred to as biotypes or host races, are abundant (Jaenike 1990), and novel examples are frequently discovered (e.g. Mlynarek and Heard 2018; Villacis-Perez et al. 2021). Specialization may be associated with variable amounts of genetic differentiation and reproductive compatibility between host races (Drès and Mallet 2002; Ehrlich and Murphy 1988; Harrison et al. 2022; Mitchell 1981), which makes them attractive models for exploring how the interplay of ecology and evolution shapes genetic structure within and among species (Berlocher and Feder 2002).
The evolution of host-specific insect lineages may be initiated by the physical separation of populations, for example following the acquisition of a new host species. This may result in reduced realized gene flow and facilitate adaptation to each host plant (Guldemond and Mackenzie 1994; Rice and Salt 1988). Specialized host lineages could also originate as a consequence of antagonistic pleiotropy or trade-offs regarding performance on different plants, favoring the linkage of performance and preference traits (Felsenstein 1982; Fry 2003; Futuyma and Peterson 1985; Jaenike 1978; Sandoval and Nosil 2005; Soudi et al. 2015). In either case, if different sets of alleles are responsible for adaptation to different plant species, offspring from parents specialized to different plants could experience reduced fitness due to their intermediate genotypes (Egan and Funk 2009; Thompson et al. 2019). This might promote the evolution of assortative mating (Howard 1993; Mackenzie and Guldemond 1994), thereby reinforcing reproductive isolation between populations.
A prime example of host plant-associated ecological specialization is the species complex formed by the pea aphid, Acyrthosiphon pisum (Hemiptera: Aphididae), a sap-sucking insect. Ac. pisum comprises multiple genetically distinct populations that differ in their preference for, and performance on, different legume genera (Fabaceae, Frantz et al. 2006; Peccoud et al. 2009; Simon et al. 2003; Via 1991). These host-associated populations typically also differ in the communities of facultative bacterial endosymbionts they harbor (Ferrari et al. 2012; Smith et al. 2015). In the pea aphid complex, host preference and host performance are heritable (Via 1991; Via 1999), the responsible loci seem to be linked (Hawthorne and Via 2001), and there is evidence for selection against both migrants and hybrids (Via et al. 2000). It appears that strong host fidelity, with individuals feeding and mating on the same plant species throughout their life cycle, provides a significant barrier to gene flow among pea aphid host races.
In contrast to Ac. pisum, a minority of aphid species are host alternating (dioecious): they undergo the sexual generation on a woody primary host plant species and most of the parthenogenetic generations on herbaceous secondary host plant species. A well-studied example of this dioecious lifestyle is the black bean aphid, Aphis fabae: females called fundatrices hatch in spring from overwintering eggs on the primary hosts (predominantly the European spindle tree, Euonymus europaeus, and the guelder-rose, Viburnum opulus). From there, their clonal offspring migrate to a large number of secondary hosts and reproduce parthenogenetically during summer (Blackman and Eastop 2000). In autumn, sexual males and females are produced and migrate back to the primary hosts, where they mate and lay overwintering eggs. Intriguingly, A. fabae also forms a complex of morphologically cryptic lineages, taxonomically treated as subspecies or species, which show a high degree of specialization to certain secondary host plant species, even though they meet and mate on common primary hosts (Blackman and Eastop 2000; Iglisch 1968; Müller 1982; Thieme 1987) (Table S1). The use of a shared mating habitat implies the potential for homogenizing gene flow among lineages, which may be counteracted by trade-offs in secondary host plant utilization (Mackenzie 1996), reduced hybrid fitness (Müller 1982; Tosh et al. 2004), or behavioral mechanisms (Raymond et al. 2001; Thieme and Dixon 1996). The understanding of the genetic structure of the A. fabae complex is still limited. While mitochondrial COI/II and CytB sequences reveal no clear genetic structure within the complex (Béji et al. 2015; Coeur d’acier et al. 2007; Coeur d’acier et al. 2014; Zhang et al. 2010), genetic differences have been found among multiple taxa using enzyme electrophoresis (Jörg and Lampel 1996). Furthermore, nuclear microsatellite markers revealed strong genetic differentiation between A. fabae cirsiiacanthoides, a taxon feeding on thistle (Cirsium vulgare) and A. fabae fabae, the nominal subspecies feeding on goosefoot (Chenopodium album) (Coeur d’acier et al. 2004; Vorburger et al. 2017). The fact that differentiation is revealed by unlinked, selectively neutral genetic markers indicates the presence of barriers to gene flow between certain members of the A. fabae complex. It suggests that, despite the use of a common mating site and the seeming lack of an environmental barrier to mating, host plant specialization of A. fabae is not (anymore) the sole result of heterogeneous selection on different summer hosts of one freely interbreeding population. However, the actual diversity of genetically diverging A. fabae lineages encountering each other on the common mating hosts remains unknown, as well as the extent of reproductive isolation among these.
The present work is based on an extensive, temporally and geographically structured dataset of A. fabae samples collected from their primary host plant E. europaeus. The samples were collected as part of a different study (Gimmi et al. 2023), which required identifying those individuals belonging to the nominal subspecies A. f. fabae by microsatellite genotyping. Here we analyzed this dataset more in depth with the goal of describing the genetic structure and diversity of A. fabae on E. europaeus. To put the original data into context, we complemented it with a collection of A. fabae individuals from the primary host plant V. opulus and from 14 different secondary host plants. We asked how many distinct genetic clusters we could identify among the collected black bean aphids, and whether individuals belonging to different genetic clusters are associated with distinct host plants. We also looked for evidence of hybridization among the distinct A. fabae lineages. Furthermore, since host race-specific endosymbiont communities are characteristic of various herbivorous insects and of the related pea aphid system in particular (Ferrari et al. 2012), we tested for the presence of the two maternally inherited, facultative bacterial endosymbionts Hamiltonella defensa and Regiella insecticola in all our aphid samples. These endosymbionts may provide different ecological functions including protection against pathogens and parasitoids (Feldhaar 2011; Guo et al. 2017; Oliver et al. 2010), but they also entail fitness costs (Polin et al. 2014; Vorburger and Gouskov 2011; Zytynska et al. 2021). Differing symbiont complements can thus be considered as an independent indication of population divergence and ecological specialization of their hosts (Ferrari et al. 2012; Hosokawa et al. 2007; Tsuchida et al. 2004).
Methods
The Aphis fabae complex
According to Blackman and Eastop (2017), the Aphis fabae complex comprises five taxa, A.f. cirsiiacanthoides, A.f. fabae, A.f. mordwilkoi, A. evonymi, and A. solanella, of which all but A.f. mordwilkoi use the European spindle tree, E. europaeus, as winter host. The guelder rose V. opulus and the mock orange Philadelphus coronarius are used as winter hosts by A.f. mordwilkoi and A.f. cirsiiacanthoides. A. evonymi does not host alternate but remains on E. europaeus throughout the year (Blackman and Eastop 2000; Lampel and Meier 2007), while the other taxa are heteroecious and use a wide range of cultivated and wild plants as summer hosts. Some of the plants are considered ‘diagnostic’ and are used to identify the different A. fabae taxa based on their acceptance of these as hosts (Müller 1982). An overview of the taxa and their host plants is presented in Table S1. Although slight morphological differences might exist between some A. fabae taxa (e.g. Müller and Steiner 1986), it is widely accepted that biological information on host plant preference should be considered to identify ‘black bean aphids’ beyond the general term A. fabae (Blackman and Eastop 2000; Heie 1986; Jörg and Lampel 1996; Lampel and Meier 2007; Müller 1982; Thieme 1987). Caution is also advised as some host plants are used by additional taxa that are not considered part of the A. fabae complex, but which are morphologically very similar (Table S1).
Aphid samples
The dataset used in this study consists of two parts: the first part comprises black bean aphids collected exclusively from their primary host E. europaeus. These samples were originally collected for a different study (Gimmi et al. 2023) in March, April, and October of the years 2019 and 2020, and in April 2021. For each time point, approximately 80 aphids were sampled in each of three municipalities near Zurich, Switzerland, situated 10 to 30 km apart from each other: Faellanden (47° 22′ N 8° 38′ E), Gossau (47° 19′ N 8° 45′ E) and Steinmaur (47° 30′ N 8° 27′ E). The three sampling sites included cultivated fields of various crops interspersed with weeds serving as summer hosts of A. fabae, and they were structured by woody hedges containing E. europaeus and V. opulus (the third possible winter host, P. coronarius, is not native to our study area). Aphids were collected within a radius of 1–2 km of the indicated sampling point depending on host plant availability. Single females were collected from host plants located at least 3 m from each other to avoid collecting aphids originating from the same clonal colony. Only wingless or visibly reproducing winged aphids were collected (virginoparae in spring and summer, gynoparae or oviparae on the winter hosts in autumn), to avoid collecting migrants that would have stopped by but not settled on the plant. The exact sample sizes and sampling dates are provided in Table S2. The second part of our sample set was collected from various host plants at multiple sites close to our research institute, including Faellanden, Gossau, and Steinmaur, and the city of Zurich (47° 22’ 0.01” N, 8° 33’ 0” E). We collected individuals from the alternative winter host V. opulus in April 2020 and April 2021, and we collected individuals from the following summer hosts in summer 2021: Achillea millefolium, Aegopodium podagraria, Anthriscus sylvestris, Arctium lappa, Beta vulgaris, Capsella bursa-pastoris, Chenopodium album, Cirsium vulgare, Cirsium arvense, Galium aparine, Galium mollugo, Matricaria chamomilla, Papaver rhoeas, Rumex obtusifolius and Tropaeolum majus. Again, samples were collected based on host plant availability but always in at least two of the municipalities Faellanden, Gossau, Steinmaur, and Zurich, taking a single female aphid per host plant individual from host plants located at least 3 m from each other. Sample sizes ranged from 14 to 37 per summer host plant species (Table S2).
DNA extraction and genotyping
Aphid DNA was extracted using a salting out protocol as in Sunnucks and Hales (1996). Each sample was genotyped at eight microsatellite loci (Af85, Af181, Af86, Af48, Af82, Afbeta, AfF, and Af50) using the primers of Coeur d’acier et al. (2004), which in previous studies showed no evidence of tight physical linkage (Sandrock et al. 2011) and proved to be reliable and successful in separating A. f. fabae and A. f. cirsiiacanthoides (Vorburger et al. 2017). Primer sequences and the PCR protocol are provided in Table S3. After PCR amplification, the microsatellite fragments were run on an ABI 3730 automated sequencer. GeneMarker 3.0.1 (SoftGenetics) was used to score the alleles. Samples were used for further analysis if the alleles of at least seven of the eight markers were successfully scored (1.4% missing data in the final dataset). In the original dataset, 16 aphid genotypes occurred twice and one genotype three times, and we kept only one sample of each genotype for further analysis. To help identify genetic clusters within our sample collection, we complemented our dataset with the genotypes of 30 samples that were clearly identified as either A. f. fabae or A. f. cirsiiacanthoides in Vorburger et al. (2017). The final dataset comprised 1619 aphid genotypes from E. europaeus and 480 from other host plants, i.e. 2099 genotypes in total (Table S2). Allele numbers per locus varied from seven to 58 (Table S6).
Analysis of genetic structure
All analyses using R were performed in Rstudio 2022.02.3 (RStudio Team 2020) with R 4.2.3 (R Core Team 2019) and using ggplot2 3.3.5 (Wickham 2016) for plotting. To assess the genetic structure present in our data and to assign samples to genetic clusters, we considered the results of three different clustering methods: snapclust (Beugin et al. 2018) implemented in the R package adegenet 2.1.5 (Jombart 2008), STRUCTURE 2.3.4 (Falush et al. 2003; Pritchard et al. 2000), and DAPC (Jombart et al. 2010) implemented in adegenet as well. Snapclust applies a combination of geometric and model-based steps and the Expectation-Maximization algorithm to cluster genotypes (Beugin et al. 2018) and runs much faster than STRUCTURE, which uses a Bayesian MCMC approach. Both rely on population genetic models assuming Hardy-Weinberg equilibrium (HWE) and linkage equilibrium within real clusters to calculate the likelihood of specific clustering solutions. DAPC is a model-free approach where the genotype data is first transformed using PCA, and then the principal components (PCs) are used as input for linear discriminant analysis (DA). As the three clustering methods yielded similar results, we only present the snapclust and STRUCTURE analyses here; details regarding the DAPC analysis can be found in the Supplementary (Analysis S1). For all clustering approaches, we arbitrarily assigned samples to a group if they showed a group membership probability >0.8.
We applied snapclust with default settings for numbers of genetic clusters (K) ranging from 1 to 20 and consulted the three information criteria AIC, BIC, and KIC to decide on the most probable K. The snapclust analysis suggested using a K value of 6 (see Results). However, the number of individuals assigned to the smallest cluster in this solution was more than 10× smaller than the number of individuals assigned to the largest cluster (Table S5), and uneven sample sizes can hamper the ‘correct’ identification of clusters (Kalinowski 2011; Neophytou 2014; Puechmaille 2016; Wang 2017). To break the influence the numerically dominant cluster might have on the detection of smaller clusters, we additionally ran snapclust on a more balanced subset of our data containing all samples from clusters 2–6 but only 222 samples from the largest cluster 1 (222 = mean number of samples in clusters 2–6). These samples consisted of the 15 A. f. fabae reference samples plus 207 samples selected randomly from those assigned to cluster 1 under K = 6.
We ran STRUCTURE with the admixture model and without prior information on sample origin. We used the settings suggested by Wang (2017) to improve detection of clusters in (possibly) unbalanced datasets. These settings include uncorrelated allele frequencies among populations (FREQSCORR = 0) and separate alpha values per population (POPALPHAS = 1, UNIFPRIORALPHA = 0), with an initial alpha of 0.17 (=1/6, six being the number of clusters inferred with snapclust). The other settings were left to their default. We ran ten independent simulations for each K between 1 and 10, doing 200,000 iterations after discarding the first 25,000 iterations as burn-in. We also ran STRUCTURE with the same settings on the more balanced data subset as described above. To infer the most probable number of genetic clusters we considered mean LnP(K) (Pritchard et al. 2000) and Evanno’s DeltaK (Evanno et al. 2005) as implemented in STRUCTURE HARVESTER (Earl and vonHoldt 2011). To summarize the output of the replicate STRUCTURE runs we used CLUMPAK (Kopelman et al. 2015).
Genetic diversity, genetic differentiation, and host plant associations
To describe genetic diversity in the microsatellite dataset, we calculated the number of alleles, observed (Ho) and expected (He) heterozygosity for all microsatellite loci overall and for each of the six genetic groups inferred by STRUCTURE with adegenet 2.1.5 (Jombart 2008). We also tested for deviations from HWE overall and within the six groups using pegas 1.1 (Paradis 2010), and we calculated pairwise FST values (Weir and Cockerham 1984) between groups with the function pairwise.WCfst and 95% confidence intervals with boot.ppfst (nboots = 1000) using hierfstat 0.5–10 (Goudet 2005). To put these values in relation to genetic differentiation that may result from spatial or temporal separation, we further calculated pairwise FST values between the three sampling sites and the different sampling time points within each of the four dominant groups found on the winter host E. europaeus (clusters 1, 2, 3, and 5). Finally, we used Fisher’s exact tests with simulated p-values (number of simulations B = 2000) to test the null hypothesis of independence between the cluster to which a sample was assigned and the host plant from which it was collected. We separately tested for winter hosts and summer hosts, leaving out samples that were not assigned to any cluster according to STRUCTURE and the references samples from A. f. fabae and A. f. cirsiiacanthoides.
Hybrid detection
To identify possible hybrids in a targeted manner we used the software NewHybrids 2.0 which applies a Bayesian clustering method (Anderson and Thompson 2002). The method requires the input data to consist of just two parental populations and their offspring. We therefore looked for hybrids separately in all pairwise combinations of the six genetic groups. Each of the input datasets consisted of the genotypes that were assigned to one of the two considered clusters with a probability >0.8, plus the genotypes whose assignment probabilities were highest to one and second highest to the other considered cluster, based on the STRUCTURE analysis under K = 6. We ran NewHybrids with a burn-in of 100,000 followed by 400,000 sweeps, using uniform priors for both π and θ, and looking for F1 hybrids only (to detect backcrosses, a larger number of markers than we have would be required). We considered those samples as hybrids that showed higher membership probability to the hybrid category than to either parental category. To estimate a detection probability for our approach, we applied NewHybrids to datasets containing simulated hybrids that we obtained with the R function adegenet::hybridize. As parental genotypes we used those individuals with an assignment probability >0.8 to the clusters under consideration in the STRUCTURE analysis and with no missing data. For each of the 15 combinations of parental clusters, we simulated 100 times 20 hybrids. On each dataset, we ran NewHybrids as above but with a burn-in of just 1000 followed by 4000 sweeps. The software detected 18.3 of the 20 simulated hybrids on average (91%, Table S13). However, the number of correctly detected simulated hybrids was particularly low for the two combinations of clusters 2 and 3 (10.3/20 = 52% of simulated hybrids detected) and 2 and 5 (16.1/20 = 80%) (Table S13). In summary, NewHybrids probably underestimates the number of hybrids for these cluster combinations, while the number of hybrids might be close to the actual number for the other combinations.
To test whether the frequency of putative hybrids differed between spring (March and April sampling timepoints) and autumn (late October sampling timepoint), we used Pearson’s χ2-square tests.
Endosymbiont detection
In many herbivorous insect species and very prominently in the pea aphid, Ac. pisum, specialized host races are characterized by carrying differing complements of heritable endosymbionts (Ferrari et al. 2012). Since the endosymbionts provide various ecological functions to their hosts and are inherited from one generation to the next, differential endosymbiont prevalences among host taxa corroborate their ecological distinctiveness. In A. fabae, all aphid individuals carry the obligate endosymbiont Buchnera aphidicola (Douglas 1998). In addition, the facultative endosymbionts Hamiltonella defensa and Regiella insecticola occur frequently, while other known facultative endosymbionts are exceedingly rare (Gimmi et al. 2023). Therefore, to test whether also in A. fabae genetically differing groups show distinct endosymbiont prevalences, we determined the presence or absence of the obligate symbiont B. aphidicola (as a positive control) and of H. defensa and R. insecticola in all our aphid samples using diagnostic PCR. We did separate PCR reactions using specific primers for each endosymbiont and determined the presence or absence of amplified endosymbiont DNA using a QIAxcel capillary electrophoresis device. The PCR protocol and primer sequences are provided in Table S4. For the analysis, we filtered out samples with missing data (A. f. fabae and A. f. cirsiiacanthoides reference samples) or negative results for B. aphidicola, remaining with N = 2047 samples. We then tested for differences in the frequency of symbiotypes (Ham-Reg-, Ham-Reg+, Ham+Reg- or Ham+Reg+) among genetic groups using pairwise Fisher’s exact tests and a Bonferroni-adjusted significance level.
Results
Genetic structure and host plant associations in the black bean aphid complex
When comparing observed heterozygosity (Ho) with expected heterozygosity (He) in our complete microsatellite dataset, we see the heterozygote deficit and significant deviation from Hardy-Weinberg equilibrium (HWE) expected for a dataset containing strong genetic structure (p < 0.0001 for all loci, Table S6).
Indeed, all clustering approaches we used point at there being multiple genetically divergent clusters in our dataset. Using snapclust, the information criteria suggest using a K value between 6 (BIC) and 7 or 8 (KIC and AIC, Fig. S1). Under K = 6, the reference samples get assigned to one cluster each and only 23 out of 2099 samples show a membership probability of less than 0.8 to any cluster. Under K = 7 or K = 8, many samples result as admixed between the new clusters, and the reference samples of A. f. fabae get assigned to two or three different clusters under K = 7 and K = 8, respectively (Fig. S3). Both these observations point at K = 6 being the best solution with snapclust. This is supported by all three information criteria suggesting K = 6 when snapclust is applied to the more balanced data subset (Fig. S2).
While LnP(K) was not informative to infer K from the STRUCTURE output, Evanno’s DeltaK suggests K = 2 (Fig. S4), which in light of the snapclust results seems overly conservative. Running STRUCTURE on the more balanced dataset, there remains some uncertainty applying Evanno’s DeltaK due to inconsistent runs at K = 5, but the posterior probabilities clearly plateau at K = 6 (Fig. S5). We therefore decided to settle with K = 6 for the following analyses. The major difference between snapclust and STRUCTURE under K = 6 is the generally lower membership probabilities resulting from the latter, resulting in more samples that are not assigned to any cluster considering STRUCTURE (147 vs. 23 with snapclust, Fig. 1). Cluster assignment based on STRUCTURE is thus more restrictive, which is why we used it for the presented follow-up analyses.
Clustering results under K = 6 from snapclust and STRUCTURE. Each aphid individual is represented by a vertical bar, the proportion of this bar in a specific color represents the likelihood that the sample belongs to the respective cluster (membership probability). For each K, the wide boxes to the left show all 2099 samples used in the analysis. The samples are ordered according to the cluster for which they show highest membership probability in the snapclust K = 6 result (x-axis). The two narrow boxes to the right zoom in on the reference samples known to represent A. f. fabae and A. f. cirsiiacanthoides, respectively.
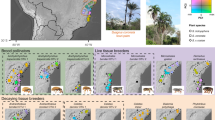
Once the samples are clustered into six genetic groups, Ho and He are close to each other within the groups, and with the exceptions of one locus each in groups 1, 2, 4, and 6, there are no significant deviations from HWE (Table S6). There is a significant correlation between the genetic group to which aphid samples are assigned and the host plant from which they were collected, both considering winter hosts (p < 0.001 in Fisher’s exact test) and summer hosts (p < 0.001, Figs. 2, S6, Tables S7–S10).
Aphids were assigned to a cluster if they showed a membership probability >0.8 to it in the STRUCTURE results under K = 6. Samples that have a membership probability below 0.8 for all clusters are categorized as "undetermined." The number of aphid individuals considered per host plant is given in brackets. Each aphid individual was collected from a different plant individual. Note that the sample size is much larger for E. euonymus than for the other host plants. a Winter host plants; b summer host plants in alphabetical order.
Among the six genetic clusters, cluster 1 likely corresponds to A. f. fabae, as it contains the respective references samples and is the only cluster found associated with A. f. fabae’s diagnostic summer host plants Beta vulgaris and Ch. album. Samples assigned to cluster 1 were collected also from the summer hosts An. sylvestris, G. aparine, M. chamomilla, P. rhoeas and R. obtusifolius (Table S9). A. f. fabae was dominant on the winter host E. europaeus (880 of 1619 samples) but rare on the winter host V. opulus (3 of 96 samples, Table S7, Fig. 2).
Cluster 2 likely corresponds to A. f. cirsiiacanthoides, as it contains the respective reference samples and is the dominant cluster found associated with Cirsium spp., A. f. cirsiiacanthoides’ diagnostic summer hosts. It was also found on several other summer hosts, including Ac. millefolium, An. sylvestris, Ar. lappa, Ca. bursa-pastoris, M. chamomilla, P. rhoeas, and R. obtusifolius. Aphids belonging to cluster 2 occurred on both winter hosts in similar frequencies (7% of all samples for both, Tables S7 and S9).
Cluster 6 likely corresponds to A. f. mordwilkoi, as it was dominant on this subspecies’ diagnostic summer hosts Ar. lappa and T. majus. Cluster 6 was also dominant on Ac. millefolium, Ae. podagraria, An. sylvestris, G. aparine and G. mollugo, and single individuals were collected from C. bursa-pastoris, Cirsium spp., M. chamomilla, and P. rhoeas. A. f. mordwilkoi was almost absent from E. europaeus (6 of 1619 samples) but frequent on V. opulus (37 of 96).
The remaining three clusters are less straightforward to identify, and we refer to the Discussion for their possible assignment to known taxa. Both cluster 5 and cluster 3 seem to use only E. europaeus as winter host (Fig. 2, Table S7). In summer, we collected aphids assigned to cluster 5 mostly from G. aparine and R. obtusifolius, while cluster 3 was virtually absent from the summer hosts we sampled (one sample on G. aparine and two on R. obtusifolius, Table S9, Fig. 2). Cluster 4 was mostly collected from V. opulus as primary host (36/38 samples) and was not observed on any of the sampled secondary host plants.
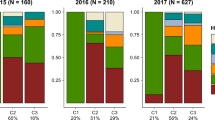
The pairwise FST values between the six genetic clusters are all significantly larger than zero, but the extent of genetic differentiation varies (Table 1). A. f. fabae and cluster 4 are most strongly differentiated from all other clusters with pairwise FST values ranging from 0.094 to 0.128 and 0.093 to 0.128, respectively. The remaining four groups are more closely related to each other with pairwise FST values between 0.050 and 0.070 (Table 1). The large number of samples collected from E. europaeus at three distinct sites and at different time points allow us to put these values in relation to genetic differentiation that may result from spatial or temporal separation. The relative proportions of the four genetic clusters dominating on E. europaeus showed some variation across space and time (Fig. 3), but within these groups, genetic differentiation was very weak, with FST values between sites or between time points vastly smaller than those between genetic clusters, and with confidence intervals that included zero in the majority of comparisons (Table S11, S12).
Distribution of individuals belonging to the six genetic clusters defined by STRUCTURE on the winter host E. euonymus between sites (Faellanden, Gossau, and Steinmaur) and over time. The four main genetic clusters (yellow, orange, violet, and blue) are present at all sites at all but one time point.
Evidence for hybridization between taxa
73 samples were inferred to be putative hybrids (Table S14, S15); 71 of them were among the 143 samples not assigned to any cluster in the main STRUCTURE analysis (‘undetermined’ samples). The putative hybrids show to the largest part admixture between clusters 1 and 4 (37 samples) and between clusters 2 and 5 (20 samples). The 1 × 4 hybrids have all been collected in spring over all sampling years and from both E. europaeus and V. opulus (32/1165 = 3% of E. europaeus spring samples, 5/91 = 5% of V. opulus spring samples). As expected for hybrid genotypes, Ho (0.80) is distinctly larger than He (0.66) within these putative 1 × 4 hybrids, and allele distributions are intermediate between those of cluster 1 and cluster 4 (Fig. S7). The putative 2 × 5 hybrids have all been collected from E. europaeus spread over all three sampling years, 18 samples in March and April and two in October. For these putative hybrids, allele distributions are intermediate between those of cluster 2 and cluster 5 (Fig. S8), but there is hardly any difference between Ho (0.70) and He (0.69). For all other pairs of clusters, we found between zero and three putative hybrids (Tables S14 and S15). Overall, 67 putative hybrids were collected in spring months (5% of spring samples from the winter hosts E. europaeus and V. opulus) and two in autumn (0.4% of autumn samples from E. europaeus). Hybrids were thus more frequent in spring than in autumn (χ2 = 19.6, df = 1, p < 0.0001). The remaining four hybrids were collected from summer host plants, one each from An. sylvestris and Ci. vulgare (hybrids of clusters 2 and 6), T. majus and P. rhoeas (hybrids of clusters 5 and 6).
Endosymbiont prevalence in Aphis fabae genetic clusters
The genetic clusters we identified with microsatellite genotypes exhibit significant differences in the prevalence of the two endosymbionts Hamiltonella defensa and Regiella insecticola (Fig. 4, Tables S16, S17). In cluster 1 (A. f. fabae) we found H. defensa in 34% and R. insecticola in 8% of the aphids. Cluster 3 shows a lower H. defensa (14%) and a much higher R. insecticola frequency (92%), while in cluster 4, 100% of the aphids carried H. defensa but only 3% R. insecticola. In the remaining three clusters, both endosymbionts are very rare (Fig. 4, Table S16). Accordingly, the symbiotypes of clusters 2, 5, and 6 do not significantly differ from each other, but they all differ from the three groups with higher endosymbiont prevalence (Fig. 4, Table S17).
Frequencies of the two endosymbiotic bacteria species Hamiltonella defensa and Regiella insecticola in each of the six genetic clusters of A. fabae, assigning samples based on the STRUCTURE results. Different letters indicate significant differences in symbiotypes in pairwise comparisons using Fisher’s exact tests with Bonferroni corrections (see Table S17 for p-values). The error bars indicate binomial proportion confidence intervals.
Among the 37 putative hybrids between clusters 1 and 4, 41% carry H. defensa and 5% R. insecticola. Statistically, symbiotypes prevalence in the 1 × 4 hybrids was not different from the prevalence in cluster 1 (p = 0.780 in Fisher’s Exact test) but different from the prevalence in cluster 4 (p < 0.000 in Fisher’s Exact test). Among the putative 2 × 5 hybrids, no sample carried H. defensa and one R. insecticola, which does not differ from the findings for cluster 2 or 5 (Tables S16, S17).
Discussion
Different plant species may impose divergent selection on the phytophagous insects exploiting them, but for the evolution and maintenance of genetically differentiated host races, reduced gene flow between host-associated populations is necessary. Here we show that despite a shared mating habitat, black bean aphids of the A. fabae complex can be assigned to at least six genetically distinct groups, which differ in their host associations and in the frequency of infection with facultative endosymbionts.
It is uncontested that such ecological diversification in phytophagous insects is facilitated when the same host plant represents adult and larval food source as well as mating site (Bush 1975; Caillaud and Via 2000; Drès and Mallet 2002; Feder et al. 1994), but our example shows that such tight linkage is not a strict requirement. A similar argument has been made for the Lepidoptera (butterflies and moths), where there is ample evidence for larval host plant-associated diversification (Braby and Trueman 2006; Braga et al. 2018; Fordyce 2010), even though adults use different food sources (often nectar) and typically mate off the larval food plants, sometimes even forming mating aggregations on hilltops (e.g. Prieto and Dahners 2009). It is noteworthy, though, that a link between adult and larval food sources may still exist in the Lepidoptera, since nectar-feeding adults include the larval food plant in their diet more often than expected by chance (Altermatt and Pearse 2011). In the A. fabae complex, a permanent link of adult and larval feeding sites with mating sites exists only for those taxa that are not host-alternating.
The existence of multiple A. fabae taxa characterized by distinct feeding preferences has been described already 100 years ago (Börner and Janisch 1922). However, the assignment of individuals to taxa using host plant choice tests may hinge on the developmental stage and the condition of both host plants and aphids (Thieme 1987), as well as on the degree of phenotypic plasticity in aphid performance traits (Gorur et al. 2005; Gorur et al. 2007). Still, in accordance with and extending on previous population genetic studies (Jörg and Lampel 1996; Vorburger et al. 2017), our data shows that there is clear genetic differentiation between black bean aphids that dominate on plants considered ’diagnostic,’ and thus between ecologically defined taxa. This attests to the careful work of the entomologists who studied this complex group with biological assays. It is particularly interesting considering the lack of resolution in mitochondrial COI/II and CytB sequences (Béji et al. 2015; Coeur d’acier et al. 2007; Coeur d’acier et al. 2014; Zhang et al. 2010), which suggests that the A. fabae lineages represent evolutionarily young taxa.
Most authors agree that the winter host E. europaeus is principally used by four A. fabae taxa (summarized by Blackman and Eastop 2017, see also Table S1). This matches well with the four main genetic groups (1, 2, 3, and 5) we found on E. europaeus, two of which we can clearly identify as A. f. fabae and A. f. cirsiiacanthoides. The other two taxa expected on E. europaeus are A. solanella, whose diagnostic summer host is Solanum nigrum, and A. evonymi, which is monoecious and feeds on E. europaeus throughout the season. As we do not have samples from either diagnostic summer host, we cannot unambiguously assign the clusters we found to the two taxa. However, we propose that cluster 3 might be either A. solanella or A. evonymi (we found samples assigned to cluster 3 almost exclusively on E. europaeus), while cluster 5 might be A. solanella but not A. evonymi, as we found it in relevant numbers on the (non-diagnostic) summer host plant R. obtusifolius and also on Galium species and P. rhoeas (Fig. 2, Table S9). Two arguments might challenge these hypotheses: first, A. evonymi is generally thought to differ visually from other A. fabae taxa due to its brownish body coloration (Blackman and Eastop 2000), but no such divergent body color was recorded during sample collection (E. Gimmi: personal observation). Second, both A. evonymi and A. solanella are considered independent species, which stands in contrast to our finding that clusters 3 and 5 are closely related to each other and to A. f. cirsiiacanthoides and A. f. mordwilkoi (Table 1). Nevertheless, because coloring is variable and the taxonomy not fully resolved, we suppose that cluster 3 is A. evonymi, and cluster 5 is A. solanella. Considering FST values in isolation, one could argue that cluster 1, cluster 4, and the clusters 2, 3, 5, and 6 together correspond to three different species or taxa, while clusters 2, 3, 5, and 6 correspond to subspecies or subtaxa (Table 1). A larger number of genetic markers, if not a genome-wide sequencing, would be useful to test this hypothesis.
On V. opulus, we could confirm the presence of two taxa expected to use this shrub as primary host according to the literature: A. f. mordwilkoi (cluster 6 – identified by its summer host associations) and A. f. cirsiiacanthoides. We did not expect to find yet another very abundant cluster that appears to be V. opulus-specific (cluster 4, green). It could either represent a yet undocumented A. fabae host race, or a different but closely related aphid species that we mistook for A. fabae when identifying aphids only by the unaided eye in the field. With hindsight, we suspect cluster 4 to represent A. viburni, a monoecious taxon that feeds on V. opulus throughout the year and is generally regarded as a member of the A. fabae complex in the broad sense (Blackman and Eastop 2000). A. viburni would show morphological differences to other black bean aphids under microscopic examination (Lampel and Meier 2007), but since we extracted DNA destructively for this study, we would need to collect new aphid samples to confirm our hypothesis. According to Coeur d’acier et al. (2007; 2014), mitochondrial markers cannot distinguish between A. viburni and members of A. fabae s. str. This is compatible with our finding that the nuclear genetic differentiation of cluster 4 from other clusters is comparable to that of A. f. fabae (Table 1) also under the assumption that cluster 4 corresponds to A. viburni.
We here confirm that there is genetic differentiation and thus restricted genetic exchange between black bean aphids associated with different secondary host plants. The relatively low number of hybrids we found is additionally indicating the presence of prezygotic or early-life postzygotic barriers to gene flow. Different mechanisms might play a role: one possibility is a difference in the timing of arrival and the production of sexual morphs on the primary host plant. Such temporal separation plays an important role in the maintenance of genetically divergent lineages in another host alternating aphid, Rhopalosiphum padi (Halkett et al. 2006; Halkett et al. 2005). While we cannot exclude some variation in the timing of sexual reproduction, this mechanism is unlikely to be relevant for reproductively separating the four main A. fabae groups on E. europaeus, since all of them were present simultaneously on E. europaeus in autumn of both sampling years (Fig. 3). However, separate temporal niches might be realized at a smaller scale, for example can mating-related activities of different taxa be unequally distributed over the day (Thieme and Dixon 1996). Also, behavioral mechanisms may contribute to reproductive isolation between A. fabae taxa: there is for instance evidence that male black bean aphids are able to differentiate between female pheromones of their own and of different taxa (Raymond et al. 2001; Thieme and Dixon 1996). We could also imagine that a behavioral preference for chemical signals from the summer host plants of specialized taxa could promote assortative mating and thus reduce gene flow between lineages. Which of these mechanisms is actually relevant for reproductive isolation among taxa within the A. fabae complex remains to be tested.
Assortative mating is selected for when hybrid offspring show reduced fitness. Based on a number of crossing experiments (Iglisch 1968; Raymond et al. 2001; Thieme 1988; Tosh et al. 2004), we can assume that reproductive success might be lower for mixed-taxa pairs than for same-taxa ones, but that viable and fertile hybrid offspring are possible. For example, Raymond et al. (2001) found hybrids between A. f. fabae and A. f. mordwilkoi to be viable but to produce fewer eggs (less than a third) than pure-bred offspring from either parental taxa. In agreement with that, we identified certain individuals in our dataset as putative hybrids (Tables S14, S15). The fact that most of these putative hybrids were collected in spring rather than in autumn is suggestive for selection acting against hybrids during the summer months, thereby reinforcing reproductive isolation between genetic groups (Howard 1993). Postzygotic selection may have an intrinsic (e.g. genetic incompatibility of parental chromosomes) or extrinsic basis, and the latter can directly be related to ecological speciation models: extrinsic postzygotic selection may manifest specifically in the environments that parental individuals are adapted to if the intermediate allele composition present in hybrids results in a reduced fitness compared to adapted parents (‘maladaptive intermediacy’, Hatfield and Schluter 1999; Rundle and Whitlock 2001). While it is not possible from our observational data to distinguish between intrinsic and extrinsic selection against hybrids (Rundle and Whitlock 2001), clear evidence for extrinsic postzygotic selection has been shown for other phytophagous insect systems (Funk 2010; Nosil et al. 2003) and might be tested for specifically in A. fabae in future experiments.
While hybridization between taxa co-occurring on the same winter hosts could be expected, we were surprised by the relatively large number of putative hybrids between cluster 1 (A. f. fabae), using E. europaeus as primary host, and cluster 4 (presumed A. viburni), using V. opulus as primary host. We observed these hybrids only in spring and on both primary host plants. That these taxa are reproductively compatible is consistent with experimental evidence from Iglisch (1968). But how are hybrids formed when the parental taxa mate on different hosts? We hypothesize that male aphids (which we did not sample) occasionally visit the ‘wrong’ hosts when actively searching for females during the period of sexual reproduction. This would be a straightforward explanation for the winged males of A. f. fabae, but less so for males of A. viburni, which are reported to be unwinged (Heie 1986). However, E. europaeus and V. opulus are very common hedgerow plants in our sampling area, often growing with intertwined branches. It would therefore at least be feasible that stray males of either taxon could mate with egg-laying females that are already settled on the correct plant species. This might explain the presence of hybrids on both winter hosts in spring despite the strict host specificity observed for the female aphids. The vicinity of the two winter host plants might also explain why genetic differentiation among taxa using the same winter host is not different from genetic differentiation among taxa using different winter hosts (cf. Table 1).
The existence of hybrids between cluster 2 (A. f. cirsiiacanthoides) and cluster 5 is less surprising, as they both mate on E. europaeus. The comparably high number (Table S14) might be either cause or result of these clusters being little differentiated (Table 1), though for the similarly differentiated combination of clusters 2 × 3 and 2 × 6, we found just one and two hybrids, respectively (Table S14, S15). Interestingly, the two 2 × 6 hybrids were both collected from summer hosts that are used by both parental clusters (Cirsium spp. and An. sylvestris). Based on this anecdotal observation, a future experiment might test whether hybrid fitness is different on host plants that are used by both parents compared to host plants that are used by either parent. This might help us understand to what extent reduced hybrid fitness is based on extrinsic compared to intrinsic fitness effects.
The correlation between the use of specific host plants and genetic differentiation in black bean aphids, combined with performance trade-offs on these different plants (Douglas 1997; Mackenzie 1996; Müller 1982), recapitulate the situation of host specialized biotypes in the pea aphid (Peccoud et al. 2009; Via 1999). The differential prevalence of heritable endosymbiotic bacteria in host-associated taxa represents another parallel between the two systems (Ferrari et al. 2012; Simon et al. 2003). In A. fabae, the prevalences of the two heritable facultative symbionts H. defensa and R. insecticola differ markedly among the different genetic groups (Fig. 4). These frequency differences may have arisen due to drift and could thus just be a consequence of the reproductive barriers existing between taxa. However, H. defensa and R. insecticola may provide their host with various ecological benefits including protection against parasitoids or pathogens (reviewed in Guo et al. 2017), but they also entail fitness costs (Polin et al. 2014; Vorburger and Gouskov 2011; Zytynska et al. 2021). Net costs are known to vary depending on the aphid’s host plant environment (McLean et al. 2011; Sochard et al. 2019). It is therefore likely that differing costs and benefits of hosting heritable endosymbionts, and thus diverging selection, account for the large differences in symbiont prevalence between A. fabae taxa specialized on different plant species (Fig. 4). Some symbionts can even directly affect aphid performance on certain host plants (Tsuchida et al. 2004; Wagner et al. 2015). No such effect is known yet for H. defensa or R. insecticola, but this might be worth to investigate in a future experiment. In either case, the symbiont frequency differences represent additional evidence for divergent ecological selection on different host plants.
In conclusion, we illustrate an example of genetic divergence within a species complex of host-alternating aphids that correlates with the association with different host plants. Genetic divergence is also correlating with differences in the frequency of infection with facultative endosymbionts. Both is suggestive of divergent selection underlying the observed differentiation, similar to host-associated diversification in other phytophagous insects. The advantage of ecological specialization seems to be strong enough to promote the maintenance of genetic divergence despite the opportunity for gene flow at shared mating sites, and this is likely achieved via an interplay of prezygotic barriers and postzygotic selection against hybrids.
Data availability
Data and scripts generated in this study are available at Dryad Digital Repository: https://doi.org/10.5061/dryad.tx95x6b5t.
References
Altermatt F, Pearse IS (2011) Similarity and specialization of the larval versus adult diet of European Butterflies and Moths. Am Nat 178:372–382. https://doi.org/10.1086/661248
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229. https://doi.org/10.1093/genetics/160.3.1217
Béji B, Bouktila D, Mezghani-Khemakhem M, Bouhachem-Boukhris S, Makni M, Makni H (2015) Structure of the black bean aphid Aphis fabae (Hemiptera: Aphididae) complex, inferred from DNA barcoding. Afr Entom 23:321–328. https://doi.org/10.4001/003.023.0206
Berlocher S, Feder J (2002) Sympatric speciation in phytophagous insects: Moving beyond controversy? Annu Rev Entomol 47:773–815. https://doi.org/10.1146/annurev.ento.47.091201.145312
Beugin MP, Gayet T, Pontier D, Devillard S, Jombart T (2018) A fast likelihood solution to the genetic clustering problem. Methods Ecol Evol 9:1006–1016. https://doi.org/10.1111/2041-210X.12968
Blackman RL, Eastop VF (2000) Aphids on the world’s crops: an identification and information guide. John Wiley and Sons Ltd, Chichester
Blackman RL, Eastop VF (2017) Taxonomic issues. In Aphids as Crop Pests, 2nd Edition (HF Van Emden, R Harrington, eds.), pp. 1-36. CABI Publishing, Wallingford UK
Börner C, Janisch R (1922) Zur Lebensgeschichte und Bekämpfung der Schwarzen Blattläuse. Nachrichtenblatt für den Deutschen Pflanzenschutz 2:65–67.
Braby MF, Trueman JWH (2006) Evolution of larval host plant associations and adaptive radiation in pierid butterflies. J Evol Biol 19:1677–1690. https://doi.org/10.1111/j.1420-9101.2006.01109.x
Braga MP, Guimarães PR, Wheat CW, Nylin S, Janz N (2018) Unifying host-associated diversification processes using butterfly–plant networks. Nat Commun 9:5155. https://doi.org/10.1038/s41467-018-07677-x
Bush GL (1975) Sympatric speciation in phytophagous parasitic insects. In Evolutionary Strategies of Parasitic Insects and Mites (PW Price, ed), pp. 187–206. Boston, MA: Springer US
Caillaud MC, Via S (2000) Specialized feeding behavior influences both ecological specialization and assortative mating in sympatric host races of pea aphids. Am Nat 156:606–621. https://doi.org/10.1086/316991
Coeur d’Acier A, Jousselin E, Martin JF, Rasplus JY (2007) Phylogeny of the genus Aphis Linnaeus, 1758 (Homoptera: Aphididae) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol 42:598–611. https://doi.org/10.1016/j.ympev.2006.10.006
Coeur d’Acier A, Cruaud A, Artige E, Genson G, Clamens A-L, Pierre E, Hudaverdian S, Simon J-C, Jousselin E, Rasplus J-Y (2014) DNA carcoding and the associated PhylAphidB@se website for the identification of European aphids (Insecta: Hemiptera: Aphididae). PLoS One 9:e97620. https://doi.org/10.1371/journal.pone.0097620
Coeur d’acier A, Sembène M, Audiot P, Rasplus JY (2004) Polymorphic microsatellites loci in the black aphid, Aphis fabae Scopoli, 1763 (Hemiptera, Aphididae). Mol Ecol Notes 4:306–308. https://doi.org/10.1111/j.1471-8286.2004.00652.x
Dobzhansky T (1940) Speciation as a stage in evolutionary divergence. Am Nat 74:312–321. https://doi.org/10.1086/280899
Douglas AE (1997) Provenance, experience and plant utilisation by the polyphagous aphid, Aphis fabae. Entomol Exp Appl 83:161–170. https://doi.org/10.1046/j.1570-7458.1997.00168.x
Douglas AE (1998) Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. https://doi.org/10.1146/annurev.ento.43.1.17
Drès M, Mallet J (2002) Host races in plant–feeding insects and their importance in sympatric speciation. Philos Trans R Soc B: Biol Sci 357:471–492. https://doi.org/10.1098/rstb.2002.1059
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Egan SP, Funk DJ (2009) Ecologically dependent postmating isolation between sympatric host forms of Neochlamisus bebbianae leaf beetles. Proc Natl Acad Sci 106:19426–19431. https://doi.org/10.1073/pnas.0909424106
Ehrlich PR, Murphy DD (1988) Plant chemistry and host range in insect herbivores. Ecology 69:908–909. https://doi.org/10.2307/1941244
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. https://doi.org/10.1093/genetics/164.4.1567
Feder JL, Opp SB, Wlazlo B, Reynolds K, Go W, Spisak S (1994) Host fidelity is an effective premating barrier between sympatric races of the apple maggot fly. Proc Natl Acad Sci USA 91:7990–7994
Feldhaar H (2011) Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36:533–543. https://doi.org/10.1111/j.1365-2311.2011.01318.x
Felsenstein J (1982) Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution 35:124–138. https://doi.org/10.1111/j.1558-5646.1981.tb04864.x
Ferrari J, West JA, Via S, Godfray HC (2012) Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66:375–390. https://doi.org/10.1111/j.1558-5646.2011.01436.x
Fordyce JA (2010) Host shifts and evolutionary radiations of butterflies. Proc R Soc B: Biol Sci 277:3735–3743. https://doi.org/10.1098/rspb.2010.0211
Frantz A, Plantegenest M, Mieuzet L, Simon JC (2006) Ecological specialization correlates with genotypic differentiation in sympatric host-populations of the pea aphid. J Evol Biol 19:392–401. https://doi.org/10.1111/j.1420-9101.2005.01025.x
Fry JD (2003) Multilocus models of sympatric speciation: Bush versus Rice versus Felsenstein. Evolution 57:1735–1746. https://doi.org/10.1111/j.0014-3820.2003.tb00582.x
Funk DJ (2010) Does strong selection promote host specialisation and ecological speciation in insect herbivores? Evidence from Neochlamisus leaf beetles. Ecol Entomol 35:41–53. https://doi.org/10.1111/j.1365-2311.2009.01140.x
Funk DJ, Filchak KE, Feder JL (2002) Herbivorous insects: model systems for the comparative study of speciation ecology, Vol. 9. Dordrecht: Springer
Futuyma DJ, Peterson SC (1985) Genetic variation in the use of resources by insects. Annu Rev Entomol 30:217–238. https://doi.org/10.1146/annurev.en.30.010185.001245
Gimmi E, Wallisch J, Vorburger C (2023) Defensive symbiosis in the wild: Seasonal dynamics of parasitism risk and symbiont-conferred resistance. Mol Ecol 32:4063–4077. https://doi.org/10.1111/mec.16976
Gorur G, Lomonaco C, Mackenzie A (2005) Phenotypic plasticity in host-plant specialisation in Aphis fabae. Ecol Entomol 30:657–664. https://doi.org/10.1111/j.0307-6946.2005.00742.x
Gorur G, Lomonaco C, Mackenzie A (2007) Phenotypic plasticity in host choice behavior in black bean aphid, Aphis fabae (Homoptera: Aphididae). Arthropod-Plant Interact 1:187–194. https://doi.org/10.1007/s11829-007-9017-0
Goudet J (2005) Hierfstat, a package for R to compute and test hierarchical F‐statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Guldemond JA, Mackenzie A (1994) Sympatric speciation in aphids. I. Host race formation by escape from gene flow. In Individuals, Populations and Patterns in Ecology (SR Leather, AD Watt, NJ Mills, KFA Walters eds.), pp. 367–378. Andower UK: Intercept
Guo J, Hatt S, He K, Chen J, Francis F, Wang Z (2017) Nine facultative endosymbionts in aphids. A review. J Asia-Pac Entomol 20:794–801. https://doi.org/10.1016/j.aspen.2017.03.025
Halkett F, Kindlmann P, Plantegenest M, Sunnucks P, Simon J-C (2006) Temporal differentiation and spatial coexistence of sexual and facultative asexual lineages of an aphid species at mating sites. J Evol Biol 19:809–815. https://doi.org/10.1111/j.1420-9101.2005.01055.x
Halkett F, Plantegenest M, Prunier-Leterme N, Mieuzet L, Delmotte F, Simon J-C (2005) Admixed sexual and facultatively asexual aphid lineages at mating sites. Mol Ecol 14:325–336. https://doi.org/10.1111/j.1365-294X.2004.02358.x
Harrison K, Tarone AM, DeWitt T, Medina RF (2022) Predicting the occurrence of host-associated differentiation in parasitic arthropods: a quantitative literature review. Entomol Exp Appl 170:5–22. https://doi.org/10.1111/eea.13123
Hatfield T, Schluter D (1999) Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution 53:866–873. https://doi.org/10.1111/j.1558-5646.1999.tb05380.x
Hawthorne DJ, Via S (2001) Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412:904–907. https://doi.org/10.1038/35091062
Heie OE (1986) The Aphidoidea (Hemiptera) of Fennoscandia and Denmark: Brill
Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T (2007) Obligate symbiont involved in pest status of host insect. Proc R Soc B: Biol 274:1979–1984. https://doi.org/10.1098/rspb.2007.0620
Howard DJ (1993) Reinforcement: origin, dynamics, and fate of an evolutionary hypothesis. In Hybrid Zones and the Evolutionary Process (RG Harrison ed.), pp. 46–69. New York: Oxford University Press.
Iglisch I (1968) Über die Entstehung der Rassen der „Schwarzen Blattläuse” (Aphis fabae Scop. und verwandte Arten), über ihre phytopathologische Bedeutung und über die Aussichten für erfolgversprechende Bekämpfungsmaßnahmen (Homoptera: Aphididae). Anz für Schädlingskunde 43:109–109. https://doi.org/10.1007/bf02041129
Jaenike J (1978) On optimal oviposition behavior in phytophagous insects. Theor Popul Biol 14:350–356. https://doi.org/10.1016/0040-5809(78)90012-6
Jaenike J (1990) Host specialization in phytophagous insects. Annu Rev Ecol Evol Syst: 243-273. https://doi.org/10.1146/annurev.es.21.110190.001331
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. J Bioinform 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jörg E, Lampel G (1996) Enzyme electrophoretic studies on the Aphis fabae group (Hom., Aphididae). J Appl Entomol 120:7–18. https://doi.org/10.1111/j.1439-0418.1996.tb01560.x
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106:625–632. https://doi.org/10.1111/1755-0998.12512
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Lampel G, Meier W (2007) Hemiptera: Sternorrhyncha-Aphidina, Vol. 2. Neuchâtel, Switzerland: CSCF and SEG.
Mackenzie A (1996) A trade-off for host plant utilization in the black bean aphid, Aphis fabae. Evolution 50:155–162. https://doi.org/10.1111/j.1558-5646.1996.tb04482.x
Mackenzie A, Guldemond JA (1994) Sympatric speciation in aphids. II Host race formation in the face of gene flow. In Individuals, Populations and Patterns in Ecology (SR Leather, AD Watt, NJ Mills, KFA Walters eds.), pp. 379-395. Andover UK: Intercept.
Matsubayashi KW, Ohshima I, Nosil P (2010) Ecological speciation in phytophagous insects. Entomol Exp Appl 134:1–27. https://doi.org/10.1111/j.1570-7458.2009.00916.x
McLean AH, van Asch M, Ferrari J, Godfray HC (2011) Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc R Soc B: Biol 278:760–766. https://doi.org/10.1098/rspb.2010.1654
Mitchell R (1981) Insect behavior, resource exploitation, and fitness. Ann Rev Entomol 26:373–396. https://doi.org/10.1146/annurev.en.26.010181.002105
Mlynarek JJ, Heard SB (2018) Strong and complex host- and habitat-associated genetic differentiation in an apparently polyphagous leaf mining insect. Biol J Linn Soc 125:885–899. https://doi.org/10.1093/biolinnean/bly166
Müller FP (1982) Das Problem Aphis fabae. Z für Angew Entomol 94:432–446. https://doi.org/10.1111/j.1439-0418.1982.tb02591.x
Müller FP, Steiner H (1986) Morphologische Unterschiede und Variation der Geflügelten im Formenkreis Aphis fabae (Homoptera: Aphididae). Beitr Ent 36:209–215. https://doi.org/10.21248/contrib.entomol.36.2.209-215
Neophytou C (2014) Bayesian clustering analyses for genetic assignment and study of hybridization in oaks: effects of asymmetric phylogenies and asymmetric sampling schemes. Tree Genet Genomes 10:273–285. https://doi.org/10.1007/s11295-013-0680-2
Nosil P (2012) Ecological Speciation. Oxford University Press, Oxford, UK
Nosil P, Crespi BJ, Sandoval CP (2003) Reproductive isolation driven by the combined effects of ecological adaptation and reinforcement. Proc R Soc B: Biol 270:1911–1918. https://doi.org/10.1098/rspb.2003.2457
Oliver KM, Degnan PH, Burke GR, Moran NA (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. https://doi.org/10.1146/annurev-ento-112408-085305
Paradis E (2010) pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Peccoud J, Ollivier A, Plantegenest M, Simon J-C (2009) A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci USA 106:7495–7500. https://doi.org/10.1073/pnas.0811117106
Polin S, Simon JC, Outreman Y (2014) An ecological cost associated with protective symbionts of aphids. Ecol Evol 4:826–830. https://doi.org/10.1002/ece3.991
Prieto C, Dahners HW (2009) Resource utilization and environmental and spatio-temporal overlap of a hilltopping Lycaenid butterfly community in the Colombian Andes. J Ins Sci 9:16. https://doi.org/10.1673/031.009.1601
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Puechmaille SJ (2016) The program STRUCTURE does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Raymond B, Searle JB, Douglas AE (2001) On the processes shaping reproductive isolation in aphids of the Aphis fabae (Scop.) complex (Aphididae: Homoptera). Biol J Linn Soc 74:205–215. https://doi.org/10.1111/j.1095-8312.2001.tb01387.x
Rice WR (1987) Speciation via habitat specialization: the evolution of reproductive isolation as a correlated character. Evol Ecol 1:301–314. https://doi.org/10.1007/BF02071555
Rice WR, Salt GW (1988) Speciation via disruptive selection on habitat preference: experimental evidence. Am Nat 131:911–917. https://doi.org/10.1111/j.1558-5646.1985.tb00401.x
RStudio Team (2020) RStudio: Integrated Development for R. RStudio. PBC, Boston, MA
Rundle HD, Nosil P (2005) Ecological speciation. Ecol Lett 8:336–352. https://doi.org/10.1111/j.1461-0248.2004.00715.x
Rundle HD, Whitlock MC (2001) A genetic interpretation of ecologically dependent isolation. Evolution 55:198–201. https://doi.org/10.1111/j.0014-3820.2001.tb01284.x
Sandoval CP, Nosil P (2005) Counteracting selective regimes and host preference evolution in ecotypes of two species of walking-sticks. Evolution 59:2405–2413. https://doi.org/10.1111/j.0014-3820.2005.tb00950.x
Sandrock C, Razmjou J, Vorburger C (2011) Climate effects on life cycle variation and population genetic architecture of the black bean aphid, Aphis fabae. Mol Ecol 20:4165–4181. https://doi.org/10.1111/j.1365-294X.2011.05242.x
Schluter D (2000) The Ecology of Adaptive Radiation. Oxford University Press, Oxford
Schluter D (2001) Ecology and the origin of species. Trends Ecol Evol 16:372–380. https://doi.org/10.1016/s0169-5347(01)02198-x
Simon JC, Carre S, Boutin M, Prunier-Leterme N, Sabater-Mun B, Latorre A, Bournoville R (2003) Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc R Soc B: Biol 270:1703–1712. https://doi.org/10.1098/rspb.2003.2430
Smith AH, Lukasik P, O’Connor MP, Lee A, Mayo G, Drott MT, Doll S, Tuttle R, Disciullo RA, Messina A, Oliver KM, Russell JA (2015) Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol 24:1135–1149. https://doi.org/10.1111/mec.13095
Sochard C, Leclair M, Simon J-C, Outreman Y (2019) Host plant effects on the outcomes of defensive symbioses in the pea aphid complex. Evol Ecol 33:651–669. https://doi.org/10.1007/s10682-019-10005-4
Soudi S, Reinhold K, Engqvist L (2015) Host-associated divergence in sympatric host races of the leaf beetle Lochmaea capreae: implications for local adaptation and reproductive isolation. Biol J Linn Soc 116:169–182. https://doi.org/10.1111/bij.12547
Sunnucks P, Hales DF (1996) Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol Biol Evol 13(3):510–524. https://doi.org/10.1093/oxfordjournals.molbev.a025612
Thieme T (1987) Members of the complex of Aphis fabae Scop. and their host plants In Population Structure, Genetics and Taxonomy of Aphids and Thysanoptera (J Holmann, J. Pelikan, AFG Dixon, L Weisman eds.), pp. 314-323. The Hague NL: SPB Academic Publishing
Thieme T (1988) Zur Biologie von Aphis fabae mordwilkowi Börner und Janisch 1922 (Hom., Aphididae)1. J Appl Entomol 105:510–515. https://doi.org/10.1111/j.1439-0418.1988.tb00218.x
Thieme T, Dixon A (1996) Mate recognition in the Aphis fabae complex: daily rhythm of release and specificity of sex pheromones. Entomol Exp Appl 79:85–89. https://doi.org/10.1111/j.1570-7458.1996.tb00812.x
Thompson KA, Osmond MM, Schluter D (2019) Parallel genetic evolution and speciation from standing variation. Evol Lett 3:129–141. https://doi.org/10.1002/evl3.106
Tosh CR, Vamvatsikos PG, Hardie J (2004) A highly viable cross between Aphis fabae (Homoptera: Aphididae) clones with different plant preference. Env Entomol 33:1081–1087. https://doi.org/10.1603/0046-225X-33.4.1081
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989. https://doi.org/10.1126/science.1094611
Via S (1991) Specialized host plant performance of pea aphid clones is not altered by experience. Ecology 72:1420–1427. https://doi.org/10.2307/1941114
Via S (1999) Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution 53:1446–1457. https://doi.org/10.1111/j.1558-5646.1999.tb05409.x
Via S (2001) Sympatric speciation in animals: The ugly duckling grows up. Trends Ecol Evol 16:381–390. https://doi.org/10.1016/s0169-5347(01)02188-7
Via S, Bouck AC, Skillman S (2000) Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution 54:1626–1637. https://doi.org/10.1111/j.0014-3820.2000.tb00707.x
Villacis-Perez E, Snoeck S, Kurlovs AH, Clark RM, Breeuwer JA, Van Leeuwen T (2021) Adaptive divergence and post-zygotic barriers to gene flow between sympatric populations of a herbivorous mite. Commun Biol 4:1–12. https://doi.org/10.1038/s42003-021-02380-y
Vorburger C, Gouskov A (2011) Only helpful when required: a longevity cost of harbouring defensive symbionts. J Evol Biol 24:1611–1617. https://doi.org/10.1111/j.1420-9101.2011.02292.x
Vorburger C, Herzog J, Rouchet R (2017) Aphid specialization on different summer hosts is associated with strong genetic differentiation and unequal symbiont communities despite a common mating habitat. J Evol Biol 30:762–772. https://doi.org/10.1111/jeb.13040
Wagner SM, Martinez AJ, Ruan Y-M, Kim KL, Lenhart PA, Dehnel AC, Oliver KM, White JA (2015) Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol 29:1402–1410. https://doi.org/10.1111/1365-2435.12459
Wang J (2017) The computer program STRUCTURE for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour 17:981–990. https://doi.org/10.1111/1755-0998.12650
Weir BS Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 8:1358–1370. https://doi.org/10.1111/j.1558-5646.1984.tb05657.x
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Zhang H, Huang X, Jiang L, Qiao G, Zheng Z (2010) Subspecies differentiation of Aphis fabae Scopoli (Hemiptera, Aphididae) based on morphological and molecular data. Acta Zootax Sin 35:537–545
Zytynska SE, Tighiouart K, Frago E (2021) Benefits and costs of hosting facultative symbionts in plant-sucking insects: A meta-analysis. Mol Ecol 30:2483–2494. https://doi.org/10.1111/mec.15897
Acknowledgements
We would like to thank Cameron Hudson for useful suggestions and corrections on the manuscript, as well as three reviewers and the editor for constructive comments. Data produced and analyzed in this paper were generated in collaboration with the Genetic Diversity Centre (GDC), ETH Zurich. This research was funded by the Swiss National Science Foundation grant nr. 31003A_181969 to CV.
Funding
Open Access funding provided by Lib4RI – Library for the Research Institutes within the ETH Domain: Eawag, Empa, PSI & WSL.
Author information
Authors and Affiliations
Contributions
CV and EG designed the study. EG, JW, and CV carried out the fieldwork, EG and JW did the laboratory work. EG analyzed the data with inputs from CV. EG wrote the first draft of the manuscript which was edited and revised by EG and CV. All authors approved the final version for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
No approval of research ethics committees was required because experimental work was conducted with an unregulated invertebrate species.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor Marc Stift.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gimmi, E., Wallisch, J. & Vorburger, C. Ecological divergence despite common mating sites: Genotypes and symbiotypes shed light on cryptic diversity in the black bean aphid species complex. Heredity (2024). https://doi.org/10.1038/s41437-024-00687-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41437-024-00687-0