This question serves as a basic introduction to the three major genome viewers. One gene, ADAM23, will be examined using all three sites so that the reader can gain an appreciation of the subtle differences in information presented at each of these sites.
National Center for Biotechnology Information Map Viewer
The NCBI Map Viewer can be accessed from the NCBI's home page, at http://www.ncbi.nlm.nih.gov. Follow the hyperlink in the right-hand column labeled Map Viewer to go to the Map Viewer home page. NCBI provides Map Viewers for 17 organisms, including mammals, other vertebrates, plants, protozoa, fungi, and invertebrates. Select an organism from the pull-down menu, enter a search term in the text box, and press Go! The search term can be any element mapped in that genome, which in human includes gene symbol, GenBank accession number, marker name or disease name. For this example, change the Organism to human, and enter 'ADAM23'.
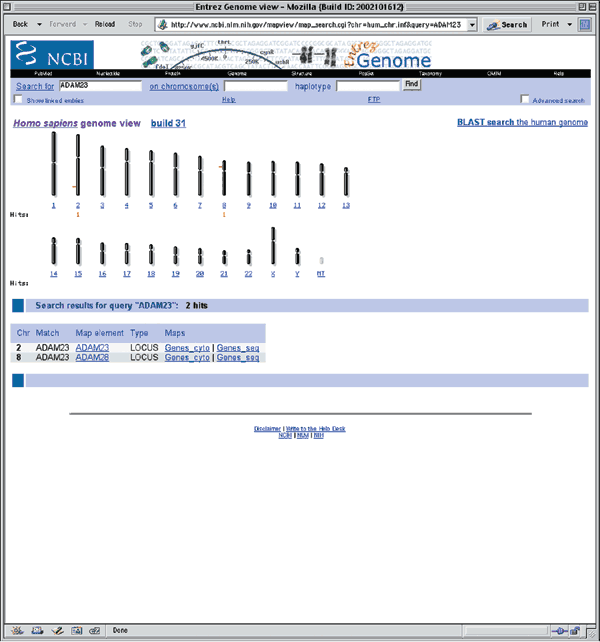
The notation at the top of the resulting page (Figure 1.1) indicates that this is Build 31, or the NCBI's 31st assembly of the human genome. Build 31 is based on sequence data from 15 November 2002. The previous genome assembly, Build 30, was based on sequence data from 28 June 2002. This overview page shows a schematic of all of the human chromosomes, pinpointing the position of ADAM23 to the q arm of chromosome 2 (Fig. 1.1). The hit on chromosome 8 is to ADAM28, which was at one time called ADAM23. The search results section shows that ADAM23 exists on two NCBI maps, Genes_cyto and Genes_seq. Genes_cyto refers to the cytogenetic map, whereas Genes_seq refers to the sequence map. Clicking on either of those two links opens a view of just that map.
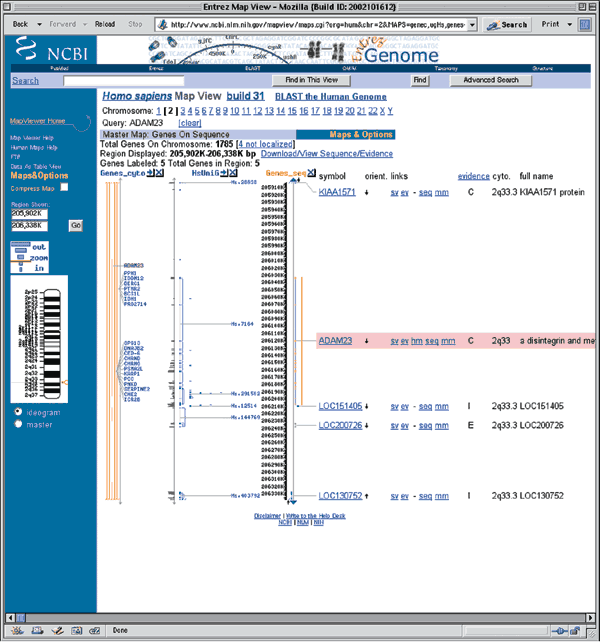
Detailed descriptions of these and other NCBI human maps are available at http://www.ncbi.nlm.nih.gov/mapview/static/humansearch.html. To get the most general overview of the genomic context of ADAM23, including all available maps, click on the item in the Map element column (in this case, ADAM23). This view shows ADAM23 and a bit of flanking sequence on chromosome 2q33. By default, the maps are shown compressed. To display the maps in a wider format, remove the check mark next to Compress Map in the left blue sidebar (Fig. 1.2). Three maps are displayed in this view, each of which will be discussed below. Additional maps, discussed in other examples in this guide, can be added to this view using the Maps & Options link.
The rightmost map is the master map, the map providing the most detail. The master map in this case is the Genes_seq map, which depicts the intron/exon organization of ADAM23 and is created by aligning the ADAM23 mRNA to the genome. The gene appears to have 27 exons. The vertical arrow next to the ADAM23 gene symbol (within the pink box) shows the direction in which the gene is transcribed. The gene symbol itself is linked to LocusLink, an NCBI resource that provides comprehensive information about the gene, including aliases, nucleotide and protein sequences, and links to other resources10 (see Question 10). The links to the right of the gene symbol point to additional information about the gene.
-
sv, or sequence view, shows the position of the gene in the context of the genomic contig, including the nucleotide and encoded protein sequences.
-
ev brings the user to the evidence viewer, a view that displays the biological evidence supporting a particular gene model. This view shows all RefSeq models, GenBank mRNAs, transcripts (whether annotated, known or potential) and expressed sequence tags (ESTs) aligning to this genomic contig. More information on the evidence viewer can be found on the NCBI Web site by clicking Evidence Viewer Help on any ev report page.
-
hm is a link to the NCBI's Human–Mouse Homology Map, showing genome sequences with predicted orthology between mouse and human (Fig. 12.2).
-
seq allows the user to retrieve the genomic sequence of the region in text format. The region of sequence displayed can easily be changed.
-
mm is a link to the Model Maker, which shows the exons that result when GenBank mRNAs, ESTs and gene predictions are aligned to the genomic sequence. The user can then select individual exons to create a customized model of the gene. More information on the Model Maker can be found on the NCBI web site by clicking help on any mm report page.
The UniG_Hs map shows human UniGene clusters that have been aligned to the genome. The gray histogram depicts the number of aligning ESTs and the blue lines show the mapping of UniGene clusters to the genome. The thick blue bars are regions of alignment (that is, exons) and the thin blue lines indicate potential introns. In this example, the mapping of UniGene cluster Hs.7164 to the genome follows that of ADAM23, and all the exons align.
The Genes_cyto map shows genes that have been mapped cytogenetically; the orange bar shows the position of the gene. Many genes have been broadly mapped to this regoin of chromosome 2.
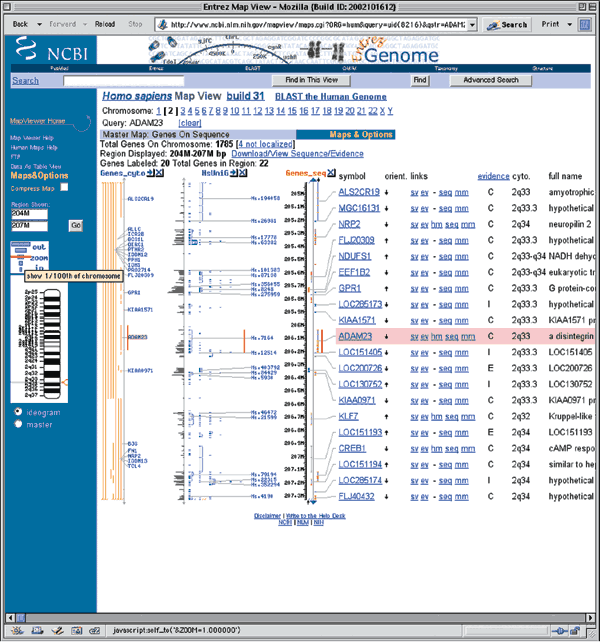
Clicking on the zoom control in the blue sidebar allows the user to zoom out to view a larger region of chromosome 2. Zooming out one level shows 1/100th of the chromosome. There are 22 genes in the region, but only 20 are labeled (displayed) in this view (Fig. 1.3). The region of ADAM23 is highlighted in red on all maps. On the basis of the Genes_seq map, ADAM23 is located between KIAA1571 and LOC151405.
University of California, Santa Cruz Genome Browser
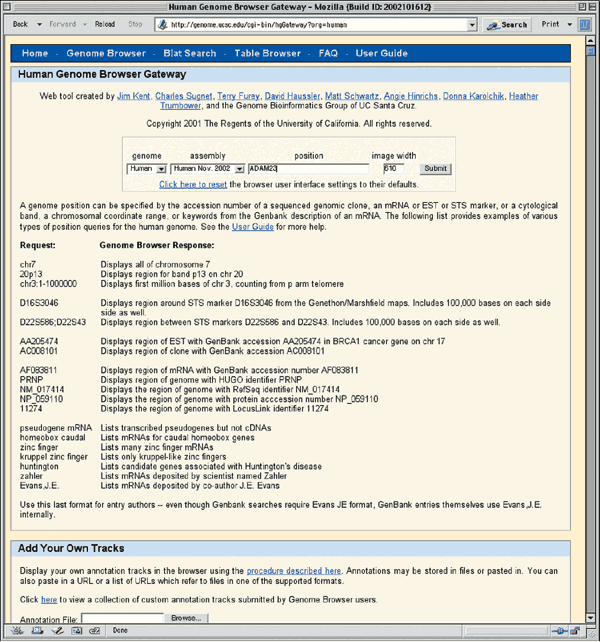
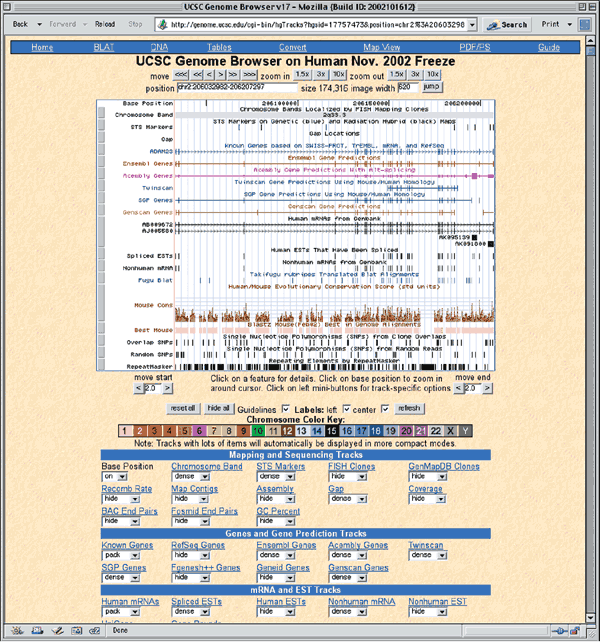
The home page for the UCSC Genome Browser is http://genome.ucsc.edu/. UCSC provides browsers not only for the most recent version of the rat, mouse, and human genome data, but also for several earlier assemblies. To use the Genome Browser, select the appropriate organism from the pull-down menu at the top of the blue sidebar (Human, in this case) and then click the link labeled Browser. On the resulting page, select the version of the human assembly to view. The Dec. 2001 browser displays annotations based on NCBI's build 28 of the human genome, the Apr. 2002 browser displays annotations on NCBI's build 29, the June 2002 browser displays annotations of NCBI's build 30, and the Nov. 2002 browser displays annotations on NCBI's build 31. Select Human Nov. 2002 from the pull-down menu to access the assembly from that date (Fig. 1.4).
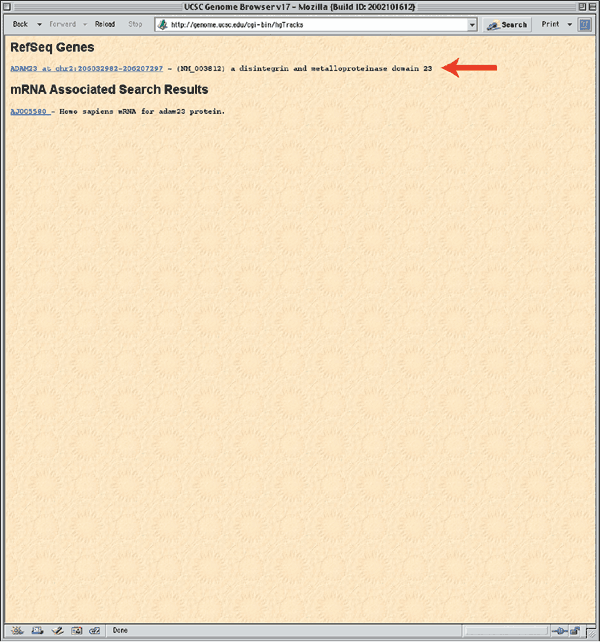
Supported types of queries are listed below the text input boxes. Enter 'ADAM23' in the box labeled position and then click Submit. The results of this search are presented in two categories, RefSeq Genes and mRNA Associated Search Results (Fig. 1.5). The section marked RefSeq Genes shows the mapping of the NCBI Reference mRNA sequences to the genome. The mRNA Associated Search Results represent the mapping of other GenBank mRNA sequences to the genome. Click on the RefSeq Genes link for ADAM23 (arrow, Fig. 1.5) to see the genomic context of the ADAM23 mRNA Reference Sequence (NM_003812).
The resulting zoomed-in view shows a region of chromosome 2 from base pair 206032982 to 206207297, located within 2q33.3 (Fig. 1.6). The blue track entitled Known Genes based on SWISS-PROT, TrEMBL, mRNA, and RefSeq shows the intron–exon structure of known genes. The vertical boxes indicate exons and the horizontal lines introns. The ADAM23 gene seems to have 26 exons. The direction of transcription is indicated by the arrowheads on the introns. The tracks labeled Ensembl Genes, Acembly Genes, Twinscan, SGP Genes, and Genscan Genes are the results of gene predictions (see Question 7). Alignments of other database nucleotide sequences are shown in the Human mRNAs from GenBank, Spliced EST, and Nonhuman mRNAs from GenBank tracks. Translated alignments of Fugu rubripes genomic sequence are in the Fugu BLAT tracks. The Mouse Cons and Best Mouse tracks shows conservation between the human and mouse genomes. Tracks displaying single-nucleotide polymorphisms (SNPs) and repetitive elements are shown at the bottom. Additional details about each track are available by selecting the track name in the Track Controls at the bottom.
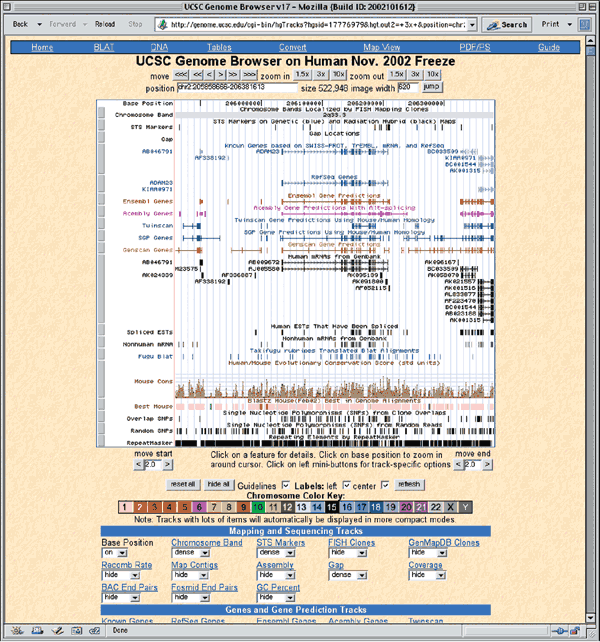
To view the genomic context of ADAM23, zoom out 3x by clicking on the zoom out 3x box in the upper right corner. ADAM23 is located between AF338192 and BC033509 (Fig. 1.7).
Ensembl
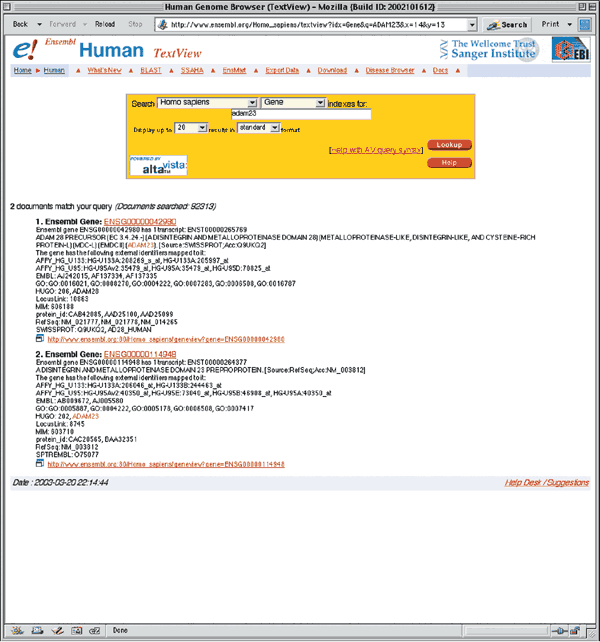
The Ensembl7 project, http://www.ensembl.org/, provides genome browsers for nine species: human, mouse, rat, zebrafish, fugu, mosquito, fruitflly, C. elegans, and C. briggsea. Click on Human to view the main entry point for the human genome. The current version of human Ensembl is version 11.31.1, based on the NCBI's 31st build of the genome. To perform a text search, enter 'ADAM23' in the text box, and limit the search by selecting Gene from the pull-down search. Click on the upper button labeled Lookup. As at NCBI, two results are returned, the first with a link to the ADAM28 gene, and the second with a link to the ADAM23 gene (Fig. 1.8).
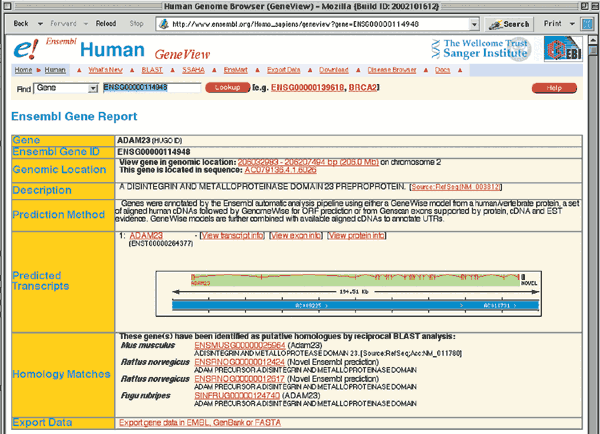
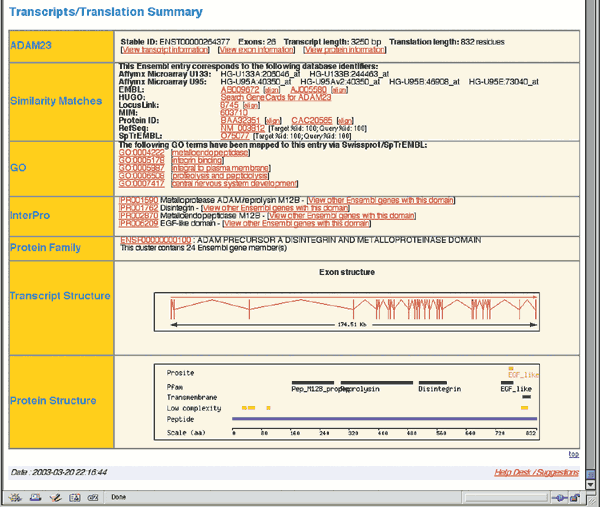
Click on either of the ADAM23 links (Ensembl Gene ENSG00000114948) to retrieve the GeneView window. The returned page contains two sections of data. The Ensembl Gene Report (Fig. 1.9) is an overview of ADAM23, including a link to the genomic location of the gene, a schematic of the intron/exon structure, and links to homologous genes from other organisms. Some of these fields will be described in more detail in later examples. The Transcripts/Translation Summary provides information on the gene transcript (Fig. 1.10). This section of the GeneView shows links to ADAM23 in other databases, as well as protein domain information. If more than one transcript is predicted for the gene, each is allocated its own summary section.
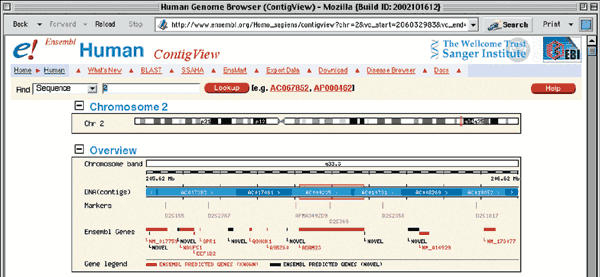
The complete genomic context of ADAM23 is viewed by returning to the first section of the GeneView (Fig. 1.9) and clicking on one of the two links within the Genomic Location box. The top portion of the resulting ContigView (Fig. 1.11) depicts the chromosome, with the region of interest outlined in red. The Overview shows the genomic context of the gene, including the chromosome bands, contigs, markers and genes that map to near 2q33.3. Clicking on any of these items recenters the display around that item. The section of interest is boxed in red on the DNA(contigs) map. The known genes annotated by Ensembl as being around ADAM23 are Q9BZ60 and NM_014929.
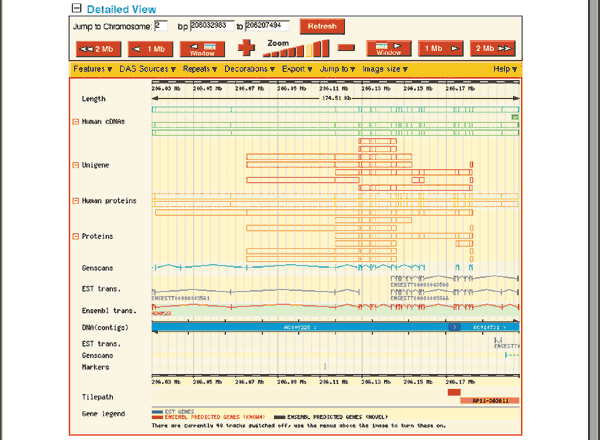
The middle panel of the ContigView, the Detailed View (Fig. 1.12), shows a zoomed-in view of the boxed region, highlighting all features that have been mapped to this region of the human genome. The navigator buttons between the Overview and the Detailed View move the display to the left and right and zoom in and out. The features to be displayed can be changed by selecting the Features pull-down menu and then checking which features to view.
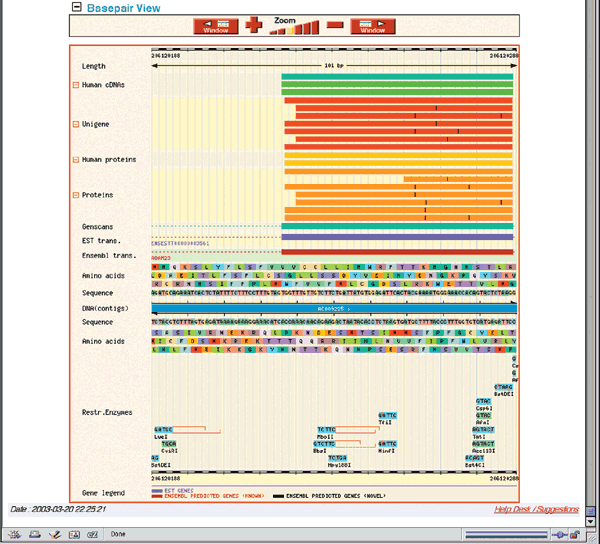
The Features shown in Fig. 1.12 are the defaults. The DNA (contigs) map separates items on the forward strand (above) from those on the reverse (below). The forward strand shows seven types of features. Starting at the bottom, the ADAM23 transcript is shown in red, indicating that it is a known transcript corresponding to a near-full-length cDNA sequence, protein sequence or both already available in the public sequence database. Black transcripts are predicted based on EST or protein sequence similarity. EST Transcr. links to individual aligning ESTs, whereas the UniGene track near the top displays UniGene clusters. The Genscan model on the forward strand contains many exons found in the known transcript and was predicted by the GENSCAN gene prediction program11 (see Question 7). The Proteins and Human proteins boxes indicate protein sequences that align to this version of the genome, whereas Human cDNAs shows mRNA sequences in the EMBL nucleotide sequence database and NCBI RefSeqs. Positioning the computer mouse over any feature brings up the feature's name and links to more detailed information. The only features on the reverse strand in this view are portions of an EST transcript and a Genscan transcript. The Basepair view, at the bottom of the ContigView (Figure 1.13) shows a very fine view of a 101 nucleotide region of ADAM23, showing the actual nucleotide and protein sequence, as well as restriction enzyme sites.
The NCBI, UCSC and Ensembl sometimes use different symbols for the same genes, so it can be difficult to compare the views obtained by the different browsers. Furthermore, the three sites maintain independent annotation pipelines and do not all attempt to align the same mRNA sequences to the genome. All three sites are currently displaying annotations based on NCBI's build 31. However, it takes significant time to update an annotation based on a new genome assembly, so shortly after the release of a new assembly, the sites may display different versions. At present, UCSC is the only site to maintain browsers based on older assemblies. Nevertheless, it is fairly easy to navigate among the three sites. The NCBI, for example, links to Ensembl and UCSC through the black boxes at the top of LocusLink entries for human genes, and Ensembl directs users to NCBI and UCSC through the “Jump to” link in its ContigView. Some versions of UCSC's Genome Browser have links to Ensembl and NCBI's Map Viewer in the blue bar at the top of each browser page.
References
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Collins, F.S. and McKusick, V.A. Implications of the Human Genome Project for medical science. J. Am. Med. Assoc. 285, 540–544 (2001).
Watson, J.D. & Crick, F.H.C. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Green, E.D. Strategies for the systematic sequencing of complex genomes. Nature Rev. Genet. 2, 573–583 (2001).
Ouellette, B.F.F. & Boguski, M.S. Database divisions and homology search files: a guide for the perplexed. Genome Res. 7, 952–955 (1997).
Bairoch, A. & Apweiler, R. The SWISS-PROT Protein Sequence Database and its supplement TREMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Hubbard, T. et al. The Ensembl Genome Database Project. Nucleic Acids Res. 30, 38–41 (2002).
Kent, W.J. BLAT—the BLAST-like Alignment Tool. Genome Res. 12, 656–664 (2002).
Stein, L. Genome annotation: from sequence to biology. Nature Rev. Genet. 2, 493–503 (2001).
Pruitt, K.D. & Maglott, D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29, 137–140 (2001).
Burge, C.B. & Karlin, S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 8, 346–354 (1998).
Schuler, G.D. Electronic PCR: bridging the gap between genome mapping and genome sequencing. Trends Biotechnol. 16, 456–459 (1998).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Hamosh, A. et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 30, 52–55 (2002).
Baxevanis, A.D. & Ouellette, B.F.F. (eds.) Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins (John Wiley & Sons, New York, 2001).
Solovyev, V.V., Salamov, A.A. & Lawrence, C.B. Identification of human gene structure using linear discriminant functions and dynamic programming. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3, 367–375 (1995).
Yeh, R.F., Lim, L.P. & Burge, C.B. Computational inference of homologous gene structures in the human genome. Genome Res. 11, 803–816 (2001).
Marchler-Bauer, A. et al. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30, 281–283 (2002).
Apweiler, R. et al. InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16, 1145–1150 (2000).
Rebhan, M., Chalifa-Caspi, V., Prilusky, J. & Lancet, D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 14, 656–664 (1998).
Blake, J.A., Richardson, J.E., Bult, C.J., Kadin, J.A. & Eppig, J.T. The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res. 30, 113–115 (2002).
Hudson, T.J. et al. A radiation hybrid map of mouse genes. Nature Genet. 29, 201–205 (2001).
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 30, 276–280 (2002).
Letunic, I. et al. Recent improvements to the SMART domain–based sequence annotation resource. Nucleic Acids Res. 30, 242–244 (2002).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Durbin, R., Eddy, S., Krogh, A. & Mitchison, G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids (Cambridge Univ. Press, Cambridge, 1998).
Peri, S., Ibarrola, N., Blagoev, B., Mann, M. & Pandey, A. Common pitfalls in bioinformatics-based analyses: look before you leap. Trends Genet. 17, 541–545 (2001) [erratum Trends Genet. 18, 218 (2002)].
Ponting, C. Issues in predicting protein function from sequence. Brief. Bioinform. 2, 19–29 (2001).
Aparicio, S.A.J.R. How to count ... human genes. Nature Genet. 25, 129–130 (2000).
Beadle, G.W. & Tatum, E.L. Genetic control of biochemical reactions in Neurospora. Proc. Natl Acad. Sci. USA 27, 499–506 (1941).
Jeffery, C.J., Bahnson, B.J., Chien, W., Ringe, D. & Petsko, G.A. Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator. Biochemistry 39, 955–964 (2000).
Wistow, G. & Piatigorsky, J. Recruitment of enzymes as lens structural proteins. Science 236, 1554–1556 (1987).
Jeffery, C.J. Moonlighting proteins. Trends Biochem. Sci. 24, 8–11 (1999).
Chothia, C. Proteins. One thousand families for the molecular biologist. Nature 357, 543–544 (1992).
Hegyi, H. & Gerstein, M. The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J. Mol. Biol. 288, 147–164 (1999).
Jansen, R. & Gerstein, M. Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res. 28, 1481–1488 (2000).
Brenner, S.E. Errors in genome annotation. Trends Genet. 15, 132–133 (1999).
Smith, R.F. Perspectives: sequence data base searching in the era of large-scale genomic sequencing. Genome Res. 6, 653–660 (1996).
Rights and permissions
About this article
Cite this article
Question 1 How does one find a gene of interest and determine that gene's structure? Once the gene has been located on the map, how does one easily examine other genes in that same region?. Nat Genet 35 (Suppl 1), 9–17 (2003). https://doi.org/10.1038/ng1189
Issue Date:
DOI: https://doi.org/10.1038/ng1189
This article is cited by
-
Cytokine signaling to the cell cycle
Immunologic Research (2007)
-
Determining to Divide: How Do Cells Decide?
Journal of Biological Physics (2005)