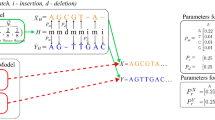
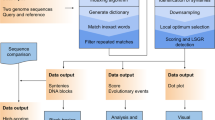
Abstract
Searching vast and rapidly growing nucleotide content in resources, such as runs in the Sequence Read Archive and assemblies for whole-genome shotgun sequencing projects in GenBank, is currently impractical for most researchers. Here we present Pebblescout, a tool that navigates such content by providing indexing and search capabilities. Indexing uses dense sampling of the sequences in the resource. Search finds subjects (runs or assemblies) that have short sequence matches to a user query, with well-defined guarantees and ranks them using informativeness of the matches. We illustrate the functionality of Pebblescout by creating eight databases that index over 3.7 petabases. The web service of Pebblescout can be reached at https://pebblescout.ncbi.nlm.nih.gov. We show that for a wide range of query lengths, Pebblescout provides a data-driven way for finding relevant subsets of large nucleotide resources, reducing the effort for downstream analysis substantially. We also show that Pebblescout results compare favorably to MetaGraph and Sourmash.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All runs and assemblies mentioned in the manuscript were publicly available at NCBI on 1 July 2023. Access to the databases is through the search functionality available on the web page https://pebblescout.ncbi.nlm.nih.gov/. Data analysis utilized Nucleotide–Nucleotide BLAST 2.14.1+, SKESA 2.5.1, SAUTE 1.3.2, SPAdes genome assembler v3.15.5 (coronaSPAdes mode), Bowtie 2 version 2.2.6, https://metagraph.ethz.ch/search and https://branchwater.sourmash.bio websites. Source data are provided with this paper.
Code availability
The metadata for all databases built (WGS, RefSeq and SRA subsets as mentioned in Table 1), queries and output for all applications presented, the software for Pebblescout and an example for building a small database and searching the built database are available at Zenodo (https://doi.org/10.5281/zenodo.10553679).
References
Sayers, E. W., O’Sullivan, C. & Karsch-Mizrachi, I. Using GenBank and SRA. Methods Mol. Biol. 2443, 1–25 (2022).
SRA database growth. NCBI https://www.ncbi.nlm.nih.gov/sra/docs/sragrowth/
GenBank and WGS statistics. NCBI https://www.ncbi.nlm.nih.gov/genbank/statistics/
Institut Pasteur project aims to index global sequencing data. Genomeweb https://www.genomeweb.com/informatics/institut-pasteur-project-aims-index-global-sequencing-data#.Y_y5nnbMI-U (2023).
Ondov, B. D. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016).
Bradley, P., den Bakker, H. C., Rocha, E. P. C., McVean, G. & Iqbal, Z. Ultrafast search of all deposited bacterial and viral genomic data. Nat. Biotechnol. 37, 152–159 (2019).
Yu, Y. et al. SeqOthello: querying RNA-seq experiments at scale. Genome Biol. 19, 167 (2018).
Holley, G. & Melsted, P. Bifrost: highly parallel construction and indexing of colored and compacted de Bruijn graphs. Genome Biol. 21, 249 (2020).
Gupta, G. et al. Fast processing and querying of 170 TB of genomics data via a Repeated And Merged BloOm Filter (RAMBO). In Proc. 2021 International Conference on Management of Data 2226–2234 (Association for Computing Machinery, 2021).
Almodaresi, F., Sarkar, H., Srivastava, A. & Patro, R. A space and time-efficient index for the compacted colored de Bruijn graph. Bioinformatics 34, i169–i177 (2018).
Pandey, P. et al. Mantis: a fast, small, and exact large-scale sequence-search index. Cell Syst. 7, 201–207.e4 (2018).
Karasikov, M. et al. MetaGraph: indexing and analysing nucleotide archives at petabase-scale. Preprint at bioRxiv https://doi.org/10.1101/2020.10.01.322164
Lemane, T., Medvedev, P., Chikhi, R. & Peterlongo, P. kmtricks: efficient and flexible construction of Bloom filters for large sequencing data collections. Bioinform. Adv. 2, vbac029 (2022).
Pierce, N. T., Irber, L., Reiter, T., Brooks, P. & Brown, C. T. Large-scale sequence comparisons with sourmash. F1000Res 8, 1006 (2019).
Karasikov, M., Mustafa, H., Rätsch, G. & Kahles, A. Lossless indexing with counting de Bruijn graphs. Genome Res. 32, 1754–1764 (2022).
Srikakulam, S. K., Keller, S., Dabbaghie, F., Bals, R. & Kalinina, O. V. MetaProFi: an ultrafast chunked Bloom filter for storing and querying protein and nucleotide sequence data for accurate identification of functionally relevant genetic variants. Bioinformatics 39, btad101 (2023).
Alanko, J. N., Vuohtoniemi, J., Mäklin, T. & Puglisi, S. J. Themisto: a scalable colored k-mer index for sensitive pseudoalignment against hundreds of thousands of bacterial genomes. Bioinformatics 39, i260–i269 (2023).
Mehringer, S. et al. Hierarchical interleaved Bloom filter: enabling ultrafast, approximate sequence queries. Genome Biol. 24, 131 (2023).
Elworth, R. A. L. et al. To petabytes and beyond: recent advances in probabilistic and signal processing algorithms and their application to metagenomics. Nucleic Acids Res. 48, 5217–5234 (2020).
Marchet, C. et al. Data structures based on k-mers for querying large collections of sequencing data sets. Genome Res. 31, 1–12 (2021).
Edgar, R. C. et al. Petabase-scale sequence alignment catalyses viral discovery. Nature 602, 142–147 (2022).
Irinyi, L., Roper, M., Malik, R. & Meyer, W. Finding a needle in a haystack—in silico search for environmental traces of Candida auris. Jpn J. Infect. Dis. 75, 490–495 (2022).
Katz, K. S. et al. STAT: a fast, scalable, MinHash-based k-mer tool to assess Sequence Read Archive next-generation sequence submissions. Genome Biol. 22, 270 (2021).
Sun, X., Kan, C., Ma, W., Du, Z. & Li, M. Genomic analysis of the suspicious SARS-CoV-2 sequences in the public sequencing database. Microbiol. Spectr.11, e0342622 (2023).
Gruber-Vodicka, H. R., Seah, B. K. B. & Pruesse, E. phyloFlash: rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems 5, e00920 (2020).
Davison, H. R., Hurst, G. D. D. & Siozios, S. ‘Candidatus Megaira’ are diverse symbionts of algae and ciliates with the potential for defensive symbiosis. Microb. Genom. 9, mgen000950 (2023).
Levi, K., Rynge, M., Abeysinghe, E., & Edwards, R. A. Searching the Sequence Read Archive using Jetstream and Wrangler. In Proc. Practice and Experience on Advanced Research Computing 1–7 (Association for Computing Machinery, 2018).
Pascar, J. & Chandler, C. H. A bioinformatics approach to identifying Wolbachia infections in arthropods. PeerJ 6, e5486 (2018).
Mori, H. et al. PZLAST: an ultra-fast amino acid sequence similarity search server against public metagenomes. Bioinformatics 37, 3944–3946 (2021).
1000 Genomes Project Consortium.A global reference for human genetic variation. Nature 526, 68–74 (2015).
Callanan, J. et al. Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci. Adv. 6, eaay5981 (2020).
Schneier, B. Description of a new variable-length key, 64-bit block cipher (Blowfish). In Proc. Fast Software Encryption, Cambridge Security Workshop 191–204 (Springer, 1993).
Schleimer, S., Wilkerson, D. S., & Aiken, A. Winnowing: local algorithms for document fingerprinting. In Proc. 2003 ACM SIGMOD International Conference on Management of Data 76–85 (Association for Computing Machinery, 2003).
Michael, R., Hayes, W., Hunt, B. R., Mount, S. M. & Yorke, J. A. Reducing storage requirements for biological sequence comparison. Bioinformatics 20, 3363–3369 (2004).
Huffman, D. A method for the construction of minimum-redundancy codes. Proc. IRE 40, 1098–1101 (1952).
Zhu, Y., Huang, W. E. & Yang, Q. Clinical perspective of antimicrobial resistance in bacteria. Infect. Drug Resist. 15, 735–746 (2022).
Becker, K. et al. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg. Infect. Dis. 24, 242–248 (2018).
Souvorov, A. & Agarwala, R. SAUTE: sequence assembly using target enrichment. BMC Bioinform. 22, 375 (2021).
Arora-Williams, K. et al. Abundant and persistent sulfur-oxidizing microbial populations are responsive to hypoxia in the Chesapeake Bay. Environ. Microbiol. 22, 2315–2332 (2022).
Gobeille, R. C. & Baskins, D. L. Data structure and storage and retrieval method supporting ordinality based searching and data retrieval. US patent US6735595B2 assigned to Hewlett Packard Enterprise Development LP; https://patents.google.com/patent/US6735595B2/en (2000).
General purpose dynamic array—Judy. Source Forge; https://sourceforge.net/projects/judy/ (2002).
Acknowledgements
This research work was supported by the NCBI of the National Library of Medicine, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank E. Yaschenko for navigating the issues related to making the Pebblescout search publicly available and to S. Ponomarov for developing the web page for the same. NCBI’s systems team, specifically R. Patterson, was very helpful in providing access to tools for monitoring system loads. We thank D. Lipman for his interest, suggestions for improving the manuscript, and introducing us to A. Fire and C. Lareau who tested and used the web page in their work, and to S. Preheim who suggested the similar metagenomic runs application. We also thank P. Ghosh, B. Robbertse and I. Tolstoy for taking an active interest as early users of Pebblescout, A. Souvorov for doing an independent assessment of alignment and assemblies for several runs, V. Schneider for providing extensive editorial comments on drafts of the manuscript and J. Asherman for his comments on readability of the manuscript from a layman’s perspective.
Author information
Authors and Affiliations
Contributions
S.A.S. proposed the presented solution and designed and implemented the software. R.A. managed the project, did testing and found applications. Both authors contributed to building databases, data interpretation and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Artem Babaian, Andre Kahles and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Lei Tang, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary dataset discussion, output formats, supplementary applications and assessments, References 41–57, Tables 1–9 and Figs. 1–5.
Source data
Source Data Fig. 2
Statistical source data.
Rights and permissions
About this article
Cite this article
Shiryev, S.A., Agarwala, R. Indexing and searching petabase-scale nucleotide resources. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02280-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41592-024-02280-z