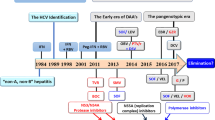
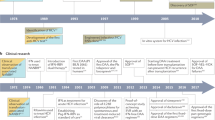
Abstract
Hepatitis C virus (HCV) will continue to be a serious global health threat for many years to come because of the chronic nature of the infection, its high prevalence and the significant morbidity of the resulting disease. Recently, a small number of molecules have produced encouraging results in proof-of-concept clinical trials. At the same time, preclinical evidence is accumulating that development of resistance will eventually limit the efficacy of new drugs. Thus, combinations of multiple agents will be required to treat chronic HCV infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wasley, A. & Alter, M. J. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20, 1–16 (2000).
Brown, R. S. Jr & Gaglio, P. J. Scope of worldwide hepatitis C problem. Liver Transpl. 9, S10–S13 (2003).
Tomei, L., Altamura, S., Paonessa, G., De Francesco, R. & Migliaccio, G. HCV antiviral resistance: the impact of in vitro studies on the development of antiviral agents targeting the viral NS5B polymerase. Antiviral Chem. Chemother. 16, 225–245 (2005).
Urbani, A. et al. Substrate specificity of the hepatitis C virus serine protease NS3. J. Biol. Chem. 272, 9204–9209 (1997).
Yan, Y. et al. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 7, 837–847 (1998).
Kim, J. L. et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87, 343–355 (1996).
Love, R. A. et al. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87, 331–342 (1996).
Llinas-Brunet, M. et al. Peptide-based inhibitors of the hepatitis C virus serine protease. Bioorg. Med. Chem. Lett. 8, 1713–1718 (1998).
Steinkuhler, C. et al. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry 37, 8899–8905 (1998).
Lamarre, D. et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426, 186–189 (2003).
Llinas-Brunet, M. et al. Highly potent and selective peptide-based inhibitors of the hepatitis C virus serine protease: towards smaller inhibitors. Bioorg. Med. Chem. Lett. 10, 2267–2270 (2000).
Goudreau, N. et al. NMR structural characterization of peptide inhibitors bound to the Hepatitis C virus NS3 protease: design of a new P2 substituent. J. Med. Chem. 47, 123–132 (2004).
Tsantrizos, Y. S. et al. Macrocyclic inhibitors of the NS3 protease as potential therapeutic agents of hepatitis C virus infection. Angew. Chem. Int. Ed. Engl. 42, 1356–1360 (2003).
Hinrichsen, H. et al. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127, 1347–1355 (2004).
Reiser, M. et al. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41, 832–835 (2005).
Lu, L. et al. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48, 2260–2266 (2004).
Lin, C. et al. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279, 17508–17514 (2004).
Trozzi, C. et al. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77, 3669–3679 (2003).
Thibeault, D. et al. Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J. Virol. 78, 7352–7359 (2004).
Chen, S. -H. et al. P1 and P1′ optimization of [3,4]-bicycloproline P2 incorporated tetrapeptidyl α-ketoamide based HCV protease inhibitors. Lett. Drug Des. Disc. 2, 118–123 (2005).
Vertex Pharmaceuticals reports that oral hepatitis C protease inhibitor VX-950 dramatically reduces viral levels in phase Ib clinical study. http://www.vrtx.com/Pressreleases2005/pr051705.html (2005).
Lemon, S. M., Yi, M. & Li, K. Strong reasons make strong actions. The antiviral efficacy of NS3/4A protease inhibitors. Hepatology 41, 671–673 (2005).
Lin, C. et al. In vitro resistance mutations against VX-950 and BILN 2061, two protease inhibitor clinical candidates: single-resistance, cross-resistance and fitness. Hepatology 40 (Suppl. 1), 404A (2004).
Behrens, S. E., Tomei, L. & De Francesco, R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15, 12–22 (1996).
Ago, H. et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7, 1417–1426 (1999).
Bressanelli, S. et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl Acad. Sci. USA 96, 13034–13049 (1999).
Lesburg, C. A. et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol. 6, 937–943 (1999).
Biswal, B. K. et al. Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280, 18202–18210 (2005).
Beaulieu, P. L. & Tsantrizos, Y. S. Inhibitors of the HCV NS5B polymerase: new hope for the treatment of hepatitis C infections. Curr. Opin. Investig. Drugs 5, 838–850 (2004).
Afdhal, N. et al. Final phase I/II trial results for NM283, a new polymerase inhibitor for hepatitis C: antiviral efficacy and tolerance in patients with HCV-1 infection, including previous interferon failures. http://www.idenix.com/products/datapres_nm283/AfdhalAASLD04_10-04.pdf (2004).
Standring, D. N. NM283 has potent antiviral activity against chronic hepatitis C virus, genotype 1, in the chimpanzee. http://www.idenix.com/products/datapres_nm283/StandringEASL2004_7-03.pdf (2004).
Carroll, S. et al. Susceptibility of different genotypes of hepatitis C virus to inhibition by nucleoside and nonnucleoside inhibitors. Antiviral Res. 62, A83 (2004).
Migliaccio, G. et al. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278, 49164–49170 (2003).
Olsen, D. B. et al. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48, 3944–3953 (2004).
Afdal, N. et al. Enhanced antiviral efficacy for Valopicitabine (NM283) plus peg-interferon in hepatitis C patients with HCV genotype-1 infection: Results of a phase IIa multicenter trial. http://www.idenix.com/products/datapres_nm283/AfdhalEASL2005_4-05.pdf (2005).
Ludmerer, S. W. et al. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49, 2059–2069 (2005).
Hashimoto, H., Mizutani, K. & Yoshida, A. in WO 00147883 (Japan Tobacco Inc., Published International Patent Application, 2001).
LaPlante, S. R. et al. Binding mode determination of benzimidazole inhibitors of the hepatitis C virus RNA polymerase by a structure and dynamics strategy. Angew Chem. Int. Ed. Engl. 43, 4306–4311 (2004).
Tomei, L. et al. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77, 13225–13231 (2003).
Di Marco, S. et al. Interdomain communication in hepatitis C virus polymerase abolished by small-molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. published online 13 June 2005.
Lu, H. in WO 2005/000308 (Rigel Pharmaceuticals, USA. Published International Patent Application, 2005).
Poor bioavailability results in insignificant clinical effects for Rigel R803 in phase I/II HCV trial. http://www.rigel.com/rigel/pr_1101094254 (2004).
ViroPharma announces data from HCV-086 proof of concept study. http://phx.corporate-ir.net/phoenix.zhtml?c=92320&p=irol-researchNewsArticle&ID=684145&highlight (2005).
Chan, L. et al. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2: tertiary amides. Bioorg. Med. Chem. Lett. 14, 797–800 (2004).
Chan, L. et al. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 1: Sulfonamides. Bioorg. Med. Chem. Lett. 14, 793–796 (2004).
Wang, M. et al. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278, 9489–9495 (2003).
Love, R. A. et al. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77, 7575–7581 (2003).
Dhanak, D. et al. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277, 38322–38327. (2002).
Gu, B. et al. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278, 16602–16607 (2003).
Tomei, L. et al. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78, 938–946 (2004).
Nguyen, T. T. et al. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47, 3525–3530 (2003).
Braasch, D. A. et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg. Med. Chem. Lett. 14, 1139–1143 (2004).
Foster, G. R. Past, present, and future hepatitis C treatments. Semin. Liver Dis. 24 (Suppl. 2), 97–104 (2004).
Kronke, J. et al. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78, 3436–3446 (2004).
Kapadia, S. B., Brideau-Andersen, A. & Chisari, F. V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl Acad Sci USA 100, 2014–2018 (2003).
Yokota, T. et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 4, 602–608 (2003).
Wilson, J. A. et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl Acad. Sci. USA 100, 2783–2788 (2003).
Randall, G., Grakoui, A. & Rice, C. M. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl Acad. Sci. USA 100, 235–240 (2003).
Song, E. et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Med. 9, 347–351 (2003).
Zender, L. et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc. Natl Acad. Sci. USA 100, 7797–7802 (2003).
McCaffrey, A. P. et al. RNA interference in adult mice. Nature 418, 38–39 (2002).
Han, J. et al. Inhibition of HCV replication in vivo by nuclease-resistant siRNAs that are targeted to the liver. Presented at 11th International Symp. Hepatitis C Virus and Related Viruses (Heidelberg, Germany 2004).
Wilson, J. A. & Richardson, C. D. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J. Virol. 79, 7050–8 (2005).
Benitec announces clinical candidate for treatment of hepatitis C. http://www.benitec.com/PRDownloads/Hepatitis%20C%20Clinical%20Candidate%20050905%20.pdf (2005).
O'Neill, L. A. TLRs: Professor Mechnikov, sit on your hat. Trends Immunol. 25, 687–693 (2004).
Takeda, K., Kaisho, T. & Akira, S. Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 (2003).
Boehme, K. W. & Compton, T. Innate sensing of viruses by toll-like receptors. J. Virol. 78, 7867–7873 (2004).
Iwasaki, A. & Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nature Immunol. 5, 987–995 (2004).
Hahn, Y. S. Subversion of immune responses by hepatitis C virus: immunomodulatory strategies beyond evasion? Curr. Opin. Immunol. 15, 443–449 (2003).
McKenna, K., Beignon, A. S. & Bhardwaj, N. Plasmacytoid dendritic cells: linking innate and adaptive immunity. J. Virol. 79, 17–27 (2005).
Schetter, C. & Vollmer, J. Toll-like receptors involved in the response to microbial pathogens: development of agonists for toll-like receptor 9. Curr. Opin. Drug Discov. Dev. 7, 204–210 (2004).
Coley reports results from phase I studies of Actilon™ for hepatitis C. http://www.coleypharma.com/coley/pr_1105025921 (2005).
Lee, J. et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc. Natl Acad. Sci. USA 100, 6646–6651 (2003).
Horsmans, Y. et al. Isatoribine a Toll-like receptor 7 agonist, significantly reduced plasma viral load in a clinical proof-of-concept study in patients with chronic hepatitis C virus. Hepatology 40 (Suppl. 1), 282A (2004).
Anadys Pharmaceuticals announces selection of ANA975 as a development candidate for front-line treatment of chronic hepatitis C. http://phx.corporate-ir.net/phoenix.zhtml?c=148908&p=irol-newsArticle&ID=575761&highlight (2004).
Hannon, G. J. RNA interference. Nature 418, 244–251 (2002).
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Manoharan, M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 8, 570–579 (2004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
G.M. and G.D.F are employees of IRBM P. Angeletti, a fully owned subsidiary of Merck and co.
Additional information
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
Rights and permissions
About this article
Cite this article
De Francesco, R., Migliaccio, G. Challenges and successes in developing new therapies for hepatitis C. Nature 436, 953–960 (2005). https://doi.org/10.1038/nature04080
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04080
This article is cited by
-
The contribution of bovines to human health against viral infections
Environmental Science and Pollution Research (2021)
-
Constitutive expression and cell-surface display of a bacterial β-mannanase in Lactobacillus plantarum
Microbial Cell Factories (2019)
-
Hepatitis C virus cell culture models: an encomium on basic research paving the road to therapy development
Medical Microbiology and Immunology (2019)
-
Optimization of drug combinations using Feedback System Control
Nature Protocols (2016)
-
Design, synthesis, and Gaussia luciferase Assay of triorganotin(IV)-based HCV inhibitors
Medicinal Chemistry Research (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.