Abstract
The identification and exploration of genetic loci that influence smoking behaviors have been conducted primarily in populations of the European ancestry. Here we report results of the first genome-wide association study meta-analysis of smoking behavior in African Americans in the Study of Tobacco in Minority Populations Genetics Consortium (n=32 389). We identified one non-coding single-nucleotide polymorphism (SNP; rs2036527[A]) on chromosome 15q25.1 associated with smoking quantity (cigarettes per day), which exceeded genome-wide significance (β=0.040, s.e.=0.007, P=1.84 × 10−8). This variant is present in the 5′-distal enhancer region of the CHRNA5 gene and defines the primary index signal reported in studies of the European ancestry. No other SNP reached genome-wide significance for smoking initiation (SI, ever vs never smoking), age of SI, or smoking cessation (SC, former vs current smoking). Informative associations that approached genome-wide significance included three modestly correlated variants, at 15q25.1 within PSMA4, CHRNA5 and CHRNA3 for smoking quantity, which are associated with a second signal previously reported in studies in European ancestry populations, and a signal represented by three SNPs in the SPOCK2 gene on chr10q22.1. The association at 15q25.1 confirms this region as an important susceptibility locus for smoking quantity in men and women of African ancestry. Larger studies will be needed to validate the suggestive loci that did not reach genome-wide significance and further elucidate the contribution of genetic variation to disparities in cigarette consumption, SC and smoking-attributable disease between African Americans and European Americans.
Similar content being viewed by others
Introduction
Smoking is influenced by genetic and environmental factors.1, 2 Genome-wide association studies (GWAS) in populations of European ancestry have identified genetic variation associated with smoking behaviors, including smoking initiation (SI), smoking quantity and smoking cessation (SC). An initial, large (n=10 995) GWAS of smoking quantity identified associations with genetic variants in the nicotinic acetylcholine receptor α5, α3 and β4 subunit cluster on chromosome 15q25.1.3 Genome-wide meta-analyses in three large consortia (n=74 053, 31 226 and 41 150) of smoking behaviors confirmed the finding at 15q25.1 and refined the association signal within the locus.4, 5, 6 Additional studies in diverse populations also have revealed independent signals in this region, suggesting multiple biologically functional variants.7, 8 This locus has also been reported as a susceptibility locus for lung cancer; however, whether this effect is independent of smoking behavior is unclear.9, 10 Additional regions have been identified for smoking quantity (CHRNB3/CHRNA6) on 8p11,4 CYP2A6 on 19q134, 6 and LOC100188947 on 10q256), SI (BDNF on 11p13)6 and SC (DBH on 9q34).6
To date, all published GWAS for smoking behaviors have been conducted in populations of European descent.11 Conducting GWAS in non-European populations, such as African ancestry populations is important because of their greater genetic diversity and population differences in disease allele frequency, linkage disequilibrium patterns and phenotype prevalence.12 For smoking behaviors, the need for GWAS in African American populations is particularly clear; African Americans, on average, initiate smoking later, smoke fewer cigarettes per day, yet are less likely to successfully quit smoking. Further, they have a higher risk of smoking-related lung cancer than many other populations.13 Ethnic differences in the clearance of nicotine, cotinine and other metabolites have been shown to contribute to the observed differences in cigarette consumption across populations, mediated in part by genetic variants in the cytochrome p450 2A6 gene.14, 15, 16
The genetic architecture of smoking-related traits is not well described in non-European ancestral groups, but there is evidence that genetic determinants have important implications for multiple addictive behaviors in populations globally.17 We established the Study of Tobacco in Minority Populations (STOMP) Genetics Consortium, which represents 13 GWAS studies of men and women of African ancestry, to search for risk loci for smoking behaviors in this population.
Materials and methods
Study description
The STOMP Genetics Consortium is comprised of the following studies: the Women's Health Initiative SNP Health Association Resource (n=8208), the African American GWAS consortia of Breast Cancer (n=5061) and Prostate Cancer (n=5556), the Candidate Gene Association Resource Consortium (including the Atherosclerosis Risk in Communities (n=2916) study, the Cleveland Family Study (n=632), the Coronary Artery Risk Development in Young Adults (n=953) study, the Jackson Heart Study (n=2145) and the Multi-Ethnic Study of Atherosclerosis (n=1646)), the Cardiovascular Health Study (n=801), the Healthy Aging in Neighborhoods across the Life Span Study (n=918), the Health ABC Study (n=1137), the Genetic Study of Atherosclerosis Risk (n=1175) and the Hypertension Genetic Epidemiology Network (n=1241). A description of each participating study as well as details regarding the measurement and collection of smoking data for each study are provided in Supplementary Materials. All studies had local Institutional Review Board approval for the present study and all participants provided written informed consent.
Smoking phenotypes
We examined four smoking phenotypes previously shown to be heritable in the African and European ancestry samples18, 19, 20, 21 and used in prior GWAS of smoking behavior.4, 5, 6 SI contrasted individuals who reported having smoked 100 cigarettes during their lifetime (ever smokers) with those who reported having smoked between 0 and 99 cigarettes during their lifetime (never smokers), consistent with the Centers for Disease Control classification.22 Among smokers, the age of SI (AOI) represented the age individuals began smoking. Some studies captured the age they first tried smoking, whereas others collected the age they began smoking regularly. As prior research suggests similar heritabilities and high genetic correlation between these phenotypes, we justified using either value in a general assessment of AOI. Similarly, for cigarettes smoked per day (CPD), some studies collected maximum CPD, whereas others collected average CPD. Longitudinal twin data suggests a high correlation between these variables over time, which supported using either value in our analyses. For studies that collected CPD as ranges, the mid-point of the interval was used as the data point; for example, individuals who reported the CPD category 0–4 were assigned a CPD value of 2. SC contrasted individuals who had quit smoking at interview (former smokers) with those who were current smokers. As relapse to smoking is highest within the first year after quitting,23 we tried to reduce misclassification by excluding smokers who quit within 1 year of interview within studies with available data. Table 1 presents distributions of smoking phenotypes across participating studies.
Genotyping and quality control
Each study performed its own genotyping using Illumina (San Diego, CA, USA) or Affymetrix GWAS arrays (Santa Clara, CA, USA). Supplementary Tables 1 and 2 present the details of the arrays, genotyping quality control procedures and sample exclusions (i.e., sex mismatch, call rate failure, relatedness, missing smoking and ancestry outliers) for each study. The quality control filters applied by each study were comparable; single-nucleotide polymorphisms (SNPs) with call rates <95% (except the Genetic Study of Atherosclerosis Risk, <90%), <1% minor allele frequency or significant (P<10−6) departure from Hardy–Weinberg equilibrium were excluded, as were individuals with excess autosomal heterozygosity, mismatch between reported and genetically determined sex, or first- or second-degree relatedness. Genome-wide imputation24 was carried out in each study using the software MACH, IMPUTE, BEAGLE or BIMBAM v0.99,25, 26, 27, 28, 29, 30, 31, 32 to infer genotypes for SNPs that were not genotyped directly on the platforms, but were genotyped on the HapMap phase 2 CEU and YRI samples.33 SNPs with imputation quality scores <0.5 were excluded.
Data analyses
Study-specific GWAS analysis. Each study conducted uniform cross-sectional analyses for each smoking phenotype using an additive genetic model. Logistic regression was used for discrete traits (SI and SC) and linear regression was used for quantitative traits (CPD and AOI). Continuous, quantitative traits were normalized by transformation to Z scores, owing to heavy tails and non-normality. Outliers were removed within each study, where abs (Z)>2. Link (Y)=Z scores were fit using ordinary least squares regression. To investigate potential sources of heterogeneity across studies, we examined the distribution of African ancestry in each cohort (Supplementary Figure 1). To account for population stratification and admixture, all studies adjusted for an appropriate number of eigenvectors3, 4, 5, 6, 7, 8, 9, 10 from a study-specific principal components analysis.34 In addition, study-specific analyses included adjustment for age and case status or study site, when appropriate. Genomic control inflation factors were computed using standard methods.35, 36
Meta-analyses of GWAS results. We performed fixed-effect meta-analysis for each smoking phenotype by computing pooled inverse-variance-weighted β-coefficients, s.e. and Z scores for each SNP.37 All GWAS results were corrected via genomic control before the meta-analysis. The study-specific lambda values utilized in this step ranged from 1.01 to 1.08 for SI (Supplementary Table 1). Heterogeneity across studies was investigated using the I2 statistic.38 The results presented herein are corrected by a second GC correction based on λ of the meta-analyses (λ<1.02). A significance threshold of P<5 × 10−8 was considered to indicate genome-wide significance. Linkage disequilibrium statistics for the largest of the STOMP cohorts (Women's Health Initiative, n=8208) were calculated using DPRIME (http://www.phs.wfubmc.edu/public/bios/gene/downloads.cfm). Linkage disequilibrium statistics for CEU and YRI were obtained from HapMap phase 2 33. Statistical power analysis was performed using QUANTO.39
Results
The meta-analysis included 32 389 genotyped men and women of African ancestry from 13 studies with sample sizes ranging from n=632 to n=8208 (Table 1). Our meta-analysis sample was 66.1% female, the mean age when smoking information was collected ranged from 35.5 to 73.4 years, and 52.7% were ever smokers. Among smokers, mean CPD ranged from 11.5 to 15.7, the mean AOI ranged from 17.3 to 23.3 years, and 44.8% were former smokers.
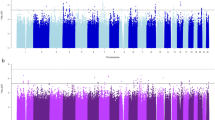
Sample sizes for the four smoking phenotype analyses (i.e., with complete genotype and phenotype data) were n=32 389 for SI, n=16 877 for AOI, n=15 547 for CPD and n=16 215 for SC. Manhattan plots for the four smoking phenotypes after double-GC scaling are shown in Figure 1. In the entire analysis, only one SNP, rs2036527, achieved genome-wide significance for one trait, CPD (β=0.04, s.e.=0.007, P=1.84 × 10−8, I2=41.6%, Table 2; study-specific results are show in Supplementary Table 3). This variant is located 6246 bp 5′ of the CHRNA5 gene on chromosome 15q25.1. We observed multiple SNPs with P-values of 10−7 associated with CPD: rs3101457, located in intron 2 (IVS2) of C1orf100 on 1q44, and rs547843, located 63 kb 5′ of a non-coding RNA sequence (LOC503519) on 15q12. Three highly correlated SNPs (r2>0.95, YRI) in the SPOCK2 gene on 10q22.1 exhibited a P-value of 10−7 with AOI (Table 2). The most significant associations for SI and SC were observed at rs566973 (∼20 kb 3′ of CRCT1 on 1q21.3) and rs3813637 (in the 3′-untranslated region of C1orf49 on 1q25.2), respectively (data not shown).
Double genomic control (GC)-corrected Manhattan plots showing significance of association of all single-nucleotide polymorphisms (SNPs) for four smoking phenotypes. (a–d). SNPs plotted on the x axis according to their position on each chromosome against, on the y axis (shown as −log10 P-value), the association with (a) smoking initiation (SI, ever vs never smokers), (b) age of SI, (c) cigarettes smoked per day, and (d) smoking cessation (former vs current smokers). Dotted red line indicates genome-wide significance threshold of P<5 × 10−8.
Four top SNPs associated with CPD span approximately 100 kb (76.6–76.7 Mb) at 15q25.1; from rs3813570, located in the 5′-untranslated region (c.-72T>C) of PSMA4, to rs938682, located in IVS4 (c.378-1941C>T) of CHRNA3 (Table 2 and Figure 2). The most significant SNP, rs2036527, is located between PSMA4 and CHRNA5, and is correlated with the index signals (rs1051730, rs16969968) for CPD reported in previous European ancestry studies. In CEU, the r2 is 0.84 between rs2036527 and rs1051730, and 0.93 between rs2036527 and rs16969968. The r2 between rs2036527 and 1051730 is 0.44 in YRI, and 0.502 in STOMP, whereas rs16969968 is non-polymorphic. Rs2036527 is also correlated with SNPs in the European Americans that tag a haplotype associated with increased expression of CHRNA5 in prefrontal cortex brain samples from European Americans and African Americans,40 but is not correlated with this haplotype in African ancestry samples (r2 between rs2036527 and rs1979905=0.443 in CEU, 0.045 in YRI and 0.064 in STOMP). The additional signals at 15q25.1 with near genome-wide significance in our study are represented by rs667282, rs938682 and rs3813570, which are weakly correlated with rs2036527 (r20.2 in CEU, 0.12 in YRI and 0.084 in STOMP). These three SNPs are correlated with each other (r20.60 in CEU and 0.32 in YRI) as well as with rs578776 and other SNPs at 15q25.1 that define a signal for smoking intensity in the European ancestry populations that is independent of rs2036527.8 However, when conditioning on rs2036527 in the four largest study populations in our sample (the African American GWAS consortia of Prostate Cancer, African American GWAS consortia of Breast Cancer, Candidate Gene Association Resource and Women's Health Initiative; n=13 113), the association between these three SNPs and CPD diminished (P-values of 10−3 after conditioning on rs2036527; Supplementary Figure 2). Assuming the GWAS arrays utilized in this study provide adequate coverage of common alleles at 15q25.1, this suggests there are not multiple independent signals for CPD in this region in African Americans or the frequencies of the functional alleles and/or their effect sizes are much smaller than the signal defined by rs2036527.
Forest and regional plot of rs2036527 with cigarettes smoked per day (CPD) from meta-analyses of the Study of Tobacco in Minority Populations (STOMP) consortia. Forest plot showing effect sizes across studies; I2=41.6%. Regional association plot show single-nucleotide polymorphisms (SNPs) plotted by position on chromosome against −log10 P-value. Estimated recombination rates (from HapMap-CEU) are plotted in light blue to reflect the local linkage disequilibrium (LD) structure on a secondary y axis. The SNPs surrounding the most significant SNP (purple) are color-coded to reflect their LD with this SNP (using pairwise r2 values from HapMap-CEU): orange, r20.8, red; 0.6–0.8, orange; 0.6–0.8; green, 0.4–0.6, light blue, 0.2–0.4; dark blue, <0.2. The blue bars at the bottom of the plot represent the relative size and location of genes in the region. AABC, African American GWAS consortia of Breast cancer; AAPC, African American GWAS consortia of Prostate Cancer; ARIC, Atherosclerosis Risk in Communities; CARDIA, Coronary Artery Risk Development in Young Adults; CFS, Cleveland Family Study; JHS, Jackson Heart Study; MESA, Multi-Ethnic Study of Atherosclerosis; HANDLS, Healthy Aging in Neighborhoods across the Life Span Study; HYPGEN, Hypertension Genetic Epidemiology Network; WHI, Women's Health Initiative.
Supplementary Table 4 presents how the variants associated with smoking behaviors in European ancestry populations performed in STOMP (rs1051730 in CHRNA3; rs16969968 in CHRNA5; rs1329650 and rs1028936 in LOC100188947; rs3733829 in EGLN2, near CYP2A6; rs6265, rs1013443, rs4923457, rs4923460, rs4074134, rs1304100, rs6484320 and rs879048 in BDNF; and rs3025343, near DBH). We observed modest nominally statistically significant associations for CPD with rs1051730 (P=0.0079) and rs16969968 (P=0.027), and for SC with rs3025343 (P=0.03).
Discussion
Investigating whether there are genetic variants associated with smoking behavior among African Americans is important, given that smoking prevalence and smoking-attributable mortality differ by race/ethnicity. Smoking prevalence and smoking intensity are lower for African Americans than European Americans, yet African Americans are less likely to successfully quit smoking.41
To our knowledge, this is the first meta-analysis of GWAS data for smoking behaviors in African Americans. The single genome-wide significant association we observed between rs2036527 and CPD is the same signal that was reported previously at 15q25.1 for nicotine dependence, smoking intensity and lung cancer in European ancestry samples.4, 5, 6, 42, 43 The strong association that we found for this SNP supports studies suggesting that it is highly correlated with the functional allele(s) in populations of African ancestry. The fact that we did not observe a strong second association signal in this region after conditioning on rs2036527 suggests that rs2036527 and correlated SNPs in the African ancestry populations may define a single common haplotype at chr15q25.1 with sufficient effect size to be detected in our sample. After back transformation of the beta estimate, mean CPD values for each rs2036527 genotype were 14.6 for AA, 13.5 for AG and 12.8 for GG, suggesting that there is an increase of less than one cigarette smoked per day for each copy of the A allele. This SNP accounted for approximately 0.20% of the phenotypic variance of CPD in our sample. This effect is similar to that reported for rs1051730, which is correlated with rs2036527, where each copy of the rs1051730 A allele corresponds to a approximately one CPD increase and accounts for 0.5% of the phenotypic variance in smoking quantity in populations of European ancestry.
A study of CHRNA5 knock-out mice showed that re-expressing this gene in the medial habenula, which extends projections to a brain region shown to mediate nicotine withdrawal,44 abolished the inhibitory effects of nicotine while maintaining the reinforcing effects of nicotine.45 In a functional magnetic resonance study of smokers, genetic variation in CHRNA5 appeared to also affect reactivity to smoking cues in the insula, hippocampus and dorsal striatum, regions implicated in addictive behavior and memory.46 Thus, it is biologically plausible that rs2036527, as a correlate of increased expression of the CHRNA5 gene, could be associated with smoking quantity as a consequence of neuro-adaptations resulting from complex interactions between genes and environment that alter positive and negative reinforcement.47
To our knowledge, no SNPs in the SPOCK2 gene, which encodes a protein that forms part of the extracellular matrix, have been reported previously in association with smoking behaviors or smoking-related cancer phenotypes. Variants at the SPOCK2 locus have been linked to bronchopulmonary dysplasia, a respiratory condition observed in premature infants48 that has been linked to intrauterine smoke exposure.49 These variants are weakly correlated with the SNPs identified at this locus for AOI in Europeans (r2<0.25 in CEU), but are not correlated in the African ancestry populations (r2=0). The top SNP associated with SC (rs3813637) is located at 1q25 in the C1orf49 gene. This locus has been linked to late-onset Alzheimer's disease, but genetic variation at this locus has not been reported in association with smoking behavior.50 We are not aware of any smoking-related, other behavioral or pathological phenotypes associated with the variants we detected at 1q44 (C1orf100) and 15q12 (LOC503519) or CTCT1 for CPD.
Although this is the largest GWAS meta-analysis of smoking phenotypes conducted to date in men and women of African ancestry, statistical power was a significant limitation. We had 80% power (for a mean allele frequency of 0.15 and α of 5 × 10−8) to detect effect sizes of 1.25 for SI, AOI and SC, and a β of 0.15 for CPD. Notably, effect sizes for variants reported with many of these smoking phenotypes reported in the larger GWAS of the European ancestry were much smaller. For example, TAG, ENGAGE and Ox-GSK consortia reported β for SI of 0.015 for SNPs in BDNF and 0.026 for rs3025343 in DBH. Thus, we cannot rule out the possibility of additional loci that influence smoking behavior among African Americans that may be detected with larger sample sizes.
This analysis was limited by the fact that we were not able to adjust for local admixture, and the chip coverage of common variants (>5%) is less complete compared with the European populations,51 which applies to most GWAS of African American populations. However, the use of a global adjustment for population genetic variation in the regression analysis using the principal components approach provided some measure of control for potential confounding because of population admixture.34, 52 Additionally, we acknowledge the limited precision of the smoking phenotypes. Smoking quantity is a highly heritable trait: estimates for CPD, heavy versus light smoking and/or pack-years range from 40 to 70% heritability in the European, African and Asian ancestry twin and family studies. Other studies have estimated that shared environmental factors account for 50% or more of the observed variation in SI, AOI and SC.1, 18, 20, 53, 54, 55, 56, 57
We were unable to directly assess more refined phenotypes and highly heritable traits such as nicotine metabolism,58 given our reliance on existing data originally collected for other purposes. Moreover, we were unable to examine gene × environment interactions using meta-GWAS analytic approach. Our analyses did not incorporate environmental covariate analyses, such as type of cigarettes smoked, mentholated or non-mentholated, dietary factors, socioeconomic status and other factors that might influence one or more of the phenotypes analyzed—data were not uniformly available and beyond the scope of the planned analyses we undertook in this discovery investigation. Future prospective studies with more detailed characterizations of smoking phenotypes and relevant environmental covariates are needed to identify additional variants that may be associated with smoking behaviors.
In summary, collective findings from GWAS among the African and European ancestry populations implicate chromosome 15q25 region as the most significant for smoking quantity. However, for both populations, SNPs in this region are associated with very small changes in smoking quantity and explain a small proportion of the variance, which suggests that conventional GWAS approaches may not be adequate to discover the likely hundreds of variants contributing small increments in risks of the additive genetic effects for heritable traits or so-called ‘missing heritability’ of complex diseases.59 The use of more refined, specific and harmonized phenotypes capturing the complex behavior of SI, trajectories of progression and cessation, and environmental effect-modifiers are also needed to detect the genetic architecture of smoking behavior in different ancestral populations. Larger studies utilizing next-generation SNP arrays, whole-exome or whole-genome sequencing will be required to investigate lower-frequency variation, which may contribute to unexplained heritability for common traits.60
References
Lessov CN, Martin NG, Statham DJ, Todorov AA, Slutske WS, Bucholz KK et al. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med 2004; 34: 865–879.
Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J . Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet 2006; 9: 64–72.
Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452: 638–642.
Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010; 42: 448–453.
Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010; 42: 436–440.
Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010; 42: 441–447.
Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D et al. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav 2010; 9: 741–750.
Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res 2009; 69: 6848–6856.
Bierut LJ . Convergence of genetic findings for nicotine dependence and smoking related diseases with chromosome 15q24-25. Trends Pharmacol Sci 2010; 31: 46–51.
Thorgeirsson TE, Stefansson K . Commentary: gene-environment interactions and smoking-related cancers. Int J Epidemiol 2010; 39: 577–579.
Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA . A Catalog of Published Genome-Wide Association Studies, Available at:##www.genome.gov/gwastudies##Accessed 25 July 2011. 2011.
Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M . Genome-wide association studies in diverse populations. Nat Rev Genet 2010; 11: 356–366.
Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med 2006; 354: 333–342.
Benowitz NL, Dains KM, Dempsey D, Wilson M, Jacob P . Racial differences in the relationship between number of cigarettes smoked and nicotine and carcinogen exposure. Nicotine Tob Res 2011; 13: 772–783.
Mwenifumbo JC, Sellers EM, Tyndale RF . Nicotine metabolism and CYP2A6 activity in a population of black African descent: impact of gender and light smoking. Drug Alcohol Depend 2007; 89: 24–33.
Moolchan ET, Berlin I, Robinson ML, Cadet JL . Characteristics of African American teenage smokers who request cessation treatment: implications for addressing health disparities. Arch Pediatr Adolesc Med 2003; 157: 533–538.
Bierut LJ . Genetic vulnerability and susceptibility to substance dependence. Neuron 2011; 69: 618–627.
Whitfield KE, King G, Moller S, Edwards CL, Nelson T, Vandenbergh D . Concordance rates for smoking among African-American twins. J Natl Med Assoc 2007; 99: 213–217.
Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT et al. A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet 2006; 79: 745–751.
Li MD, Cheng R, Ma JZ, Swan GE . A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction (Abingdon, England) 2003; 98: 23–31.
True WR, Heath AC, Scherrer JF, Waterman B, Goldberg J, Lin N et al. Genetic and environmental contributions to smoking. Addiction (Abingdon, England) 1997; 92: 1277–1287.
CDC. Cigarette smoking among adults--United States, 2007. Morb Mortal Wkly Rep 2008; 57: 1221–1226.
Hughes JR, Keely J, Naud S . Shape of the relapse curve and long-term abstinence among untreated smokers. Addiction (Abingdon, England) 2004; 99: 29–38.
Li Y, Willer C, Sanna S, Abecasis G . Genotype imputation. Annu Rev Genomics Hum Genet 2009; 10: 387–406.
Li N, Stephens M . Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 2003; 165: 2213–2233.
Howie BN, Donnelly P, Marchini J . A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Scheet P, Stephens M . A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 2006; 78: 629–644.
Browning BL, Browning SR . A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009; 84: 210–223.
Browning SR . Multilocus association mapping using variable-length Markov chains. Am J Hum Genet 2006; 78: 903–913.
Browning SR . Missing data imputation and haplotype phase inference for genome-wide association studies. Human Genet 2008; 124: 439–450.
Browning SR, Browning BL . Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81: 1084–1097.
Marchini J, Howie B . Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511.
Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet 2005; 37: 1243–1246.
Devlin B, Bennett P, Dawson G, Figlewicz DA, Grigorenko EL, McMahon W et al. Alleles of a reelin CGG repeat do not convey liability to autism in a sample from the CPEA network. Am J Med Genet B Neuropsychiatr Genet 2004; 126B: 46–50.
de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF . Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Gen 2008; 17 (R2): R122–R128.
Ioannidis JP, Patsopoulos NA, Evangelou E . Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2007; 2: e841.
Gauderman WJ, Morrison JM . QUANTO 1.1: A Computer Program for Statistical Power and Sample Size Calculations for Genetic-Epidemiology Studies 2006.
Smith RM, Alachkar H, Papp AC, Wang D, Mash DC, Wang JC et al. Nicotinic alpha5 receptor subunit mRNA expression is associated with distant 5′ upstream polymorphisms. Eur J Hum Genet 2011; 19: 76–83.
Trinidad DR, Perez-Stable EJ, White MM, Emery SL, Messer K . A nationwide analysis of US racial/ethnic disparities in smoking behaviors, smoking cessation, and cessation-related factors. Am J Public Health 2011; 101: 699–706.
Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008; 40: 616–622.
Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008; 452: 633–637.
Salas R, Sturm R, Boulter J, De Biasi M . Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J Neurosci 2009; 29: 3014–3018.
Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ . Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature 2011; 471: 597–601.
Janes AC, Smoller JW, David SP, Frederick BD, Haddad S, Basu A et al. Association between CHRNA5 genetic variation at rs16969968 and brain reactivity to smoking images in nicotine dependent women. Drug Alcohol Depend 2012; 120: 7–13.
Robinson TE, Berridge KC . The psychology and neurobiology of addiction: an incentive-sensitization view. Addiction (Abingdon, England) 2000; 95 (Suppl 2): S91–S117.
Hadchouel A, Durrmeyer X, Bouzigon E, Incitti R, Huusko J, Jarreau PH et al. Identification of SPOCK2 as a Susceptibility Gene for Bronchopulmonary Dysplasia. Am J Respir Crit Care Med 2011; 184: 1164–1170.
Antonucci R, Contu P, Porcella A, Atzeni C, Chiappe S . Intrauterine smoke exposure: a new risk factor for bronchopulmonary dysplasia? J Perinat Med 2004; 32: 272–277.
Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet 2007; 81: 17–31.
Jorgenson E, Witte JS . A gene-centric approach to genome-wide association studies. Nat Rev Genet 2006; 7: 885–891.
Patterson N, Price AL, Reich D . Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Kaprio J, Koskenvuo M, Langinvainio H . [Finnish twins reared apart. IV: smoking and drinking habits. A preliminary analysis of the effect of heredity and environment]. Acta Genet Med Gemellol 1984; 33: 425–433.
Kaprio J, Koskenvuo M, Sarna S . Cigarette smoking, use of alcohol, and leisure-time physical activity among same-sexed adult male twins. Prog Clin Biol Res 1981; 69 (Pt C): 37–46.
Lessov-Schlaggar CN, Pang Z, Swan GE, Guo Q, Wang S, Cao W et al. Heritability of cigarette smoking and alcohol use in Chinese male twins: the Qingdao twin registry. Int J Epidemiol 2006; 35: 1278–1285.
Carmelli D, Swan GE, Robinette D, Fabsitz RR . [Heritability of substance use in the NAS-NRC Twin Registry]. Acta Genet Med Gemellol (Roma) 1990; 39: 91–98.
Hettema JM, Corey LA, Kendler KS . A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend 1999; 57: 69–78.
Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL . Genetic and environmental influences on the ratio of 3′hydroxycotinine to cotinine in plasma and urine. Pharmacogenet Genomics 2009; 19: 388–398.
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al. Finding the missing heritability of complex diseases. Nature 2009; 461: 747–753.
Goldstein DB . Growth of genome screening needs debate. Nature 2011; 476: 27–28.
Acknowledgements
We wish to acknowledge the many contributors from multiple institutions and funders who contributed to this project. Detailed acknowledgements are described in the supplementary information available at Translational Psychiatry's website.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
David, S., Hamidovic, A., Chen, G. et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry 2, e119 (2012). https://doi.org/10.1038/tp.2012.41
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2012.41
Keywords
This article is cited by
-
Clarifying the Genetic Influences on Nicotine Dependence and Quantity of Use in Cigarette Smokers
Behavior Genetics (2021)
-
A decade in psychiatric GWAS research
Molecular Psychiatry (2019)
-
Association and cis-mQTL analysis of variants in CHRNA3-A5, CHRNA7, CHRNB2, and CHRNB4 in relation to nicotine dependence in a Chinese Han population
Translational Psychiatry (2018)
-
The integrated landscape of causal genes and pathways in schizophrenia
Translational Psychiatry (2018)
-
Fuzzy heaping mechanism for heaped count data with imprecision
Soft Computing (2018)