Article
|
Open Access
Featured
-
-
Article
| Open Access Single-cell analysis of chromatin accessibility in the adult mouse brain
Single-cell analysis of chromatin accessibility in the adult mouse brain
An atlas of candidate cis-regulatory DNA elements (cCREs) in the adult mouse brain unravels the transcriptional regulatory programs that drive the heterogeneity and complexity of brain structure and function.
- Songpeng Zu
- , Yang Eric Li
- & Bing Ren
-
Article
| Open Access Identification of constrained sequence elements across 239 primate genomes
Identification of constrained sequence elements across 239 primate genomes
Whole-genome alignment of 239 primate species reveals noncoding regulatory elements that are under selective constraint in primates but not in other placental mammals, that are enriched for variants that affect human gene expression and complex traits in diseases.
- Lukas F. K. Kuderna
- , Jacob C. Ulirsch
- & Kyle Kai-How Farh
-
Article |
 Complementary Alu sequences mediate enhancer–promoter selectivity
Complementary Alu sequences mediate enhancer–promoter selectivity
Using RNA in situ conformation sequencing technology, the role of Alu elements in mediating the interaction between enhancers and promoters is shown.
- Liang Liang
- , Changchang Cao
- & Yuanchao Xue
-
Article
| Open Access Cooperation between bHLH transcription factors and histones for DNA access
Cooperation between bHLH transcription factors and histones for DNA access
Cryo-EM structures and analysis provide insight into the mechanisms by which basic helix–loop–helix transcription factors access E-box DNA sequences that are embedded within nucleosomes, and cooperate with other transcription factors.
- Alicia K. Michael
- , Lisa Stoos
- & Nicolas H. Thomä
-
Article
| Open Access Histone modifications regulate pioneer transcription factor cooperativity
Histone modifications regulate pioneer transcription factor cooperativity
Binding of the human pioneer transcription factor OCT4 to nucleosomes containing endogenous DNA sequences causes changes to the nucleosome structure and facilitates the cooperative assembly of multiple pioneer transcription factors, a property that can be affected by histone modifications.
- Kalyan K. Sinha
- , Silvija Bilokapic
- & Mario Halic
-
Article |
 Mitotic bookmarking by SWI/SNF subunits
Mitotic bookmarking by SWI/SNF subunits
Subunits of SWI/SNF act as mitotic bookmarks to safeguard cell identity during cell division.
- Zhexin Zhu
- , Xiaolong Chen
- & Charles W. M. Roberts
-
Article |
 Large-scale mapping and mutagenesis of human transcriptional effector domains
Large-scale mapping and mutagenesis of human transcriptional effector domains
A high throughput recruitment assay testing the transcriptional activity of more than 100,000 protein fragments tiling across most human chromatin regulators and transcription factors maps the locations and strengths of activation, repression and bifunctional domains, and identifies the sequences necessary for these functions.
- Nicole DelRosso
- , Josh Tycko
- & Lacramioara Bintu
-
Article |
 A mechanism for oxidative damage repair at gene regulatory elements
A mechanism for oxidative damage repair at gene regulatory elements
The nuclear mitotic apparatus protein NuMA helps to protect genes from oxidative damage by occupying regions around transcription start sites, binding DNA repair factors and promoting transcription following damage.
- Swagat Ray
- , Arwa A. Abugable
- & Sherif F. El-Khamisy
-
Article |
 Differential cofactor dependencies define distinct types of human enhancers
Differential cofactor dependencies define distinct types of human enhancers
The systematic categorization of human enhancers by their cofactor dependencies provides a conceptual framework to understand the sequence and chromatin diversity of enhancers and their roles in different gene-regulatory programmes.
- Christoph Neumayr
- , Vanja Haberle
- & Alexander Stark
-
Article |
 Transcriptional coupling of distant regulatory genes in living embryos
Transcriptional coupling of distant regulatory genes in living embryos
In Drosophila, there are extensive physical and functional associations of distant paralogous genes, including co-regulation by shared enhancers and co-transcriptional initiation over distances of nearly 250 kilobases.
- Michal Levo
- , João Raimundo
- & Michael S. Levine
-
Article
| Open Access Gene regulation by gonadal hormone receptors underlies brain sex differences
Gene regulation by gonadal hormone receptors underlies brain sex differences
A study maps neuronal genomic targets of oestrogen receptor-α and shows how they coordinate brain sexual differentiation, concluding that the genome remains responsive to hormonal changes after structural dimorphisms have been established.
- B. Gegenhuber
- , M. V. Wu
- & J. Tollkuhn
-
Article
| Open Access Nonlinear control of transcription through enhancer–promoter interactions
Nonlinear control of transcription through enhancer–promoter interactions
The transcriptional effect of an enhancer depends on its contact probabilities with the promoter through a nonlinear relationship, and enhancer strength determines absolute transcription levels as well as the sensitivity of a promoter to CTCF-mediated transcriptional insulation.
- Jessica Zuin
- , Gregory Roth
- & Luca Giorgetti
-
Article |
 Enhancer release and retargeting activates disease-susceptibility genes
Enhancer release and retargeting activates disease-susceptibility genes
Disruption of a promoter can release its partner enhancer to activate other promoters in the same contact domain, and this process, named ‘enhancer release and retargeting’, can often lead to gene alterations that cause disease.
- Soohwan Oh
- , Jiaofang Shao
- & Michael G. Rosenfeld
-
Article |
 A high-resolution protein architecture of the budding yeast genome
A high-resolution protein architecture of the budding yeast genome
A ChIP–exo method is used to define the genome-wide positional organization of proteins associated with gene transcription, DNA replication, centromeres, subtelomeres and transposons, revealing distinct protein assemblies for constitutive and inducible gene expression.
- Matthew J. Rossi
- , Prashant K. Kuntala
- & B. Franklin Pugh
-
Article |
 Systematic analysis of binding of transcription factors to noncoding variants
Systematic analysis of binding of transcription factors to noncoding variants
An ultra-high-throughput multiplex protein–DNA binding assay is used to assess binding of 270 human transcription factors to 95,886 noncoding variants in the human genome, providing data to improve prediction of the effects of noncoding variants on transcription factor binding and thereby increase understanding of molecular pathways involved in diverse human traits and genetic diseases.
- Jian Yan
- , Yunjiang Qiu
- & Bing Ren
-
Article |
 DNA mismatches reveal conformational penalties in protein–DNA recognition
DNA mismatches reveal conformational penalties in protein–DNA recognition
A high-throughput assay that introduces mismatched base pairs into the DNA sequence shows that mismatches can increase transcription factor binding affinity by prepaying some of the energetic cost of distorting the DNA.
- Ariel Afek
- , Honglue Shi
- & Raluca Gordân
-
Article |
 Identification of the human DPR core promoter element using machine learning
Identification of the human DPR core promoter element using machine learning
A machine learning approach shows that the downstream core promoter region (DPR) is widely used in human gene promoters, and that many promoters contain either a DPR or a TATA box, but not both.
- Long Vo ngoc
- , Cassidy Yunjing Huang
- & James T. Kadonaga
-
Article
| Open Access The changing mouse embryo transcriptome at whole tissue and single-cell resolution
The changing mouse embryo transcriptome at whole tissue and single-cell resolution
RNA expression is quantified at a tissue level in seventeen mouse tissues across embryonic development, and at the single-cell level in the developing limb.
- Peng He
- , Brian A. Williams
- & Barbara J. Wold
-
Article |
 Nucleolar RNA polymerase II drives ribosome biogenesis
Nucleolar RNA polymerase II drives ribosome biogenesis
RNA polymerase II has an unexpected function in the nucleolus, helping to drive the expression of ribosomal RNA and to protect nucleolar structure through a mechanism involving triplex R-loop structures.
- Karan J. Abraham
- , Negin Khosraviani
- & Karim Mekhail
-
Article |
 Dynamic lineage priming is driven via direct enhancer regulation by ERK
Dynamic lineage priming is driven via direct enhancer regulation by ERK
ERK reversibly regulates embryonic stem cell transcription via selective redistribution of co-factors and RNA polymerase from pluripotency to early differentiation enhancers, while leaving transcription factors bound to their enhancers, thus preserving plasticity.
- William B. Hamilton
- , Yaron Mosesson
- & Joshua M. Brickman
-
Letter |
 BORIS promotes chromatin regulatory interactions in treatment-resistant cancer cells
BORIS promotes chromatin regulatory interactions in treatment-resistant cancer cells
The CTCF paralogue BORIS is upregulated in transcriptionally reprogrammed neuroblastoma cells rendered resistant to targeted therapy, in which it promotes regulatory chromatin interactions that maintain the resistance phenotype.
- David N. Debruyne
- , Ruben Dries
- & Rani E. George
-
Letter |
 Transcriptional cofactors display specificity for distinct types of core promoters
Transcriptional cofactors display specificity for distinct types of core promoters
A screen of 23 transcriptional cofactors for their ability to activate 72,000 candidate core promoters in Drosophila melanogaster identified distinct compatibility groups, providing insight into mechanisms that underlie the selective activation of transcriptional programs.
- Vanja Haberle
- , Cosmas D. Arnold
- & Alexander Stark
-
Letter |
 Genomic encoding of transcriptional burst kinetics
Genomic encoding of transcriptional burst kinetics
Allele-specific single-cell RNA sequencing provides insights into transcription kinetics, with data indicating that core promoter sequences affect burst size, whereas enhancers mainly affect burst frequency.
- Anton J. M. Larsson
- , Per Johnsson
- & Rickard Sandberg
-
Letter |
 A Cdk9–PP1 switch regulates the elongation–termination transition of RNA polymerase II
A Cdk9–PP1 switch regulates the elongation–termination transition of RNA polymerase II
The kinase Cdk9 and the phosphatase Dis2 regulate the termination of transcription in fission yeast in part by controlling the phosphorylation state of the elongation factor Spt5.
- Pabitra K. Parua
- , Gregory T. Booth
- & Robert P. Fisher
-
Letter |
 Chromatin analysis in human early development reveals epigenetic transition during ZGA
Chromatin analysis in human early development reveals epigenetic transition during ZGA
By applying an optimized ATAC-seq protocol to human early embryos, distinct accessible chromatin landscapes are found before and after zygotic genome activation, revealing a marked epigenetic transition during zygotic genome activation and putative regulatory elements wiring human early development.
- Jingyi Wu
- , Jiawei Xu
- & Yingpu Sun
-
Letter |
 Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer
Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer
The glycolytic enzyme PFKFB4 directly phosphorylates and regulates binding of the coactivator SRC-3 to ATF4 and thereby increases the transcriptional activity of this complex, leading to increased expression of metabolic genes, and enhancing tumour growth and metastasis.
- Subhamoy Dasgupta
- , Kimal Rajapakshe
- & Bert W. O’Malley
-
Letter |
 The cis-regulatory dynamics of embryonic development at single-cell resolution
The cis-regulatory dynamics of embryonic development at single-cell resolution
An improved assay for chromatin accessibility at single-cell resolution in Drosophila melanogaster embryos enables identification of developmental-stage- and cell-lineage-specific patterns of chromatin-level transcriptional regulation.
- Darren A. Cusanovich
- , James P. Reddington
- & Eileen E. M. Furlong
-
Letter |
 Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge
Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge
Histone deacetylase 3 (HDAC3) is required to activate brown adipose tissue enhancers to ensure thermogenic aptitude.
- Matthew J. Emmett
- , Hee-Woong Lim
- & Mitchell A. Lazar
-
Article |
 Complex multi-enhancer contacts captured by genome architecture mapping
Complex multi-enhancer contacts captured by genome architecture mapping
A technique called genome architecture mapping (GAM) involves sequencing DNA from a large number of thin nuclear cryosections to develop a map of genome organization without the limitations of existing 3C-based methods.
- Robert A. Beagrie
- , Antonio Scialdone
- & Ana Pombo
-
Article |
 An atlas of human long non-coding RNAs with accurate 5′ ends
An atlas of human long non-coding RNAs with accurate 5′ ends
A catalogue of human long non-coding RNA genes and their expression profiles across samples from major human primary cell types, tissues and cell lines.
- Chung-Chau Hon
- , Jordan A. Ramilowski
- & Alistair R. R. Forrest
-
Letter |
 Local regulation of gene expression by lncRNA promoters, transcription and splicing
Local regulation of gene expression by lncRNA promoters, transcription and splicing
Various cis-regulatory functions of genomic loci that produce long non-coding RNAs are revealed, including instances where their promoters have enhancer-like activity and the lncRNA transcripts themselves are not required for activity.
- Jesse M. Engreitz
- , Jenna E. Haines
- & Eric S. Lander
-
Letter |
 PionX sites mark the X chromosome for dosage compensation
PionX sites mark the X chromosome for dosage compensation
Recognition of the X chromosome by the dosage compensation complex in Drosophila relies on the sequence and shape of PionX sites.
- Raffaella Villa
- , Tamas Schauer
- & Peter B. Becker
-
Letter |
 The nature of mutations induced by replication–transcription collisions
The nature of mutations induced by replication–transcription collisions
When transcription and replication machineries collide on DNA, they can cause mutations to occur in the area near the collision; these mutations are now shown to include two types—duplications/deletions within the transcription unit and base substitutions in the cis-regulatory element of gene expression.
- T. Sabari Sankar
- , Brigitta D. Wastuwidyaningtyas
- & Jue D. Wang
-
Article |
 SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination
SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination
Symmetric dimethylation of the human RNA polymerase II C-terminal domain residue R1810 by the protein arginine methyltransferase 5 (PRMT5) directly recruits the protein survival of motor neuron (SMN) and indirectly recruits the helicase senataxin to resolve R-loops and promote transcription termination.
- Dorothy Yanling Zhao
- , Gerald Gish
- & Jack F. Greenblatt
-
Letter |
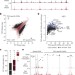 Competition between DNA methylation and transcription factors determines binding of NRF1
Competition between DNA methylation and transcription factors determines binding of NRF1
The relationship between DNA methylation and transcription factor binding was studied across the genome in mouse embryonic stem cells-the study reveals that the transcription factor NRF1 is methylation-sensitive and how physiological binding of NRF1 relies on local removal of DNA methylation.
- Silvia Domcke
- , Anaïs Flore Bardet
- & Dirk Schübeler
-
Letter |
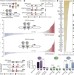 Transcriptional regulators form diverse groups with context-dependent regulatory functions
Transcriptional regulators form diverse groups with context-dependent regulatory functions
A large-scale enhancer complementation assay assessing the activating or repressing contributions of over 800 Drosophila transcription factors and cofactors to combinatorial enhancer control reveals a more complex picture than expected, with many factors having diverse regulatory functions that depend on the enhancer context.
- Gerald Stampfel
- , Tomáš Kazmar
- & Alexander Stark
-
Letter |
 DNA-dependent formation of transcription factor pairs alters their binding specificity
DNA-dependent formation of transcription factor pairs alters their binding specificity
A high-throughput analysis of DNA binding in over 9,000 interacting transcription factor pairs reveals that the interactions are often actively mediated by the DNA itself and the composite DNA sites recognized are different from the individual motifs of each transcription factor.
- Arttu Jolma
- , Yimeng Yin
- & Jussi Taipale
-
Letter |
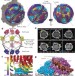 In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus
In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus
This study visualizes the interior of a dsRNA virus using cryo-electron microscopy, revealing the organization of the genome of cytoplasmic polyhedrosis virus together with its transcriptional enzyme complex in both quiescent and transcribing states.
- Xing Zhang
- , Ke Ding
- & Z. Hong Zhou
-
Article |
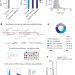 BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis
BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis
A CRISPR-Cas9 approach is used to perform saturating mutagenesis of the human and mouse BCL11A enhancers, producing a map that reveals critical regions and specific vulnerabilities; BCL11A enhancer disruption is validated by CRISPR-Cas9 as a therapeutic strategy for inducing fetal haemoglobin by applying it in both mice and primary human erythroblast cells.
- Matthew C. Canver
- , Elenoe C. Smith
- & Daniel E. Bauer
-
Letter |
 Intersecting transcription networks constrain gene regulatory evolution
Intersecting transcription networks constrain gene regulatory evolution
Epistatic interactions, whereby a mutation's effect is contingent on another mutation, have been shown to constrain evolution within single proteins, and how such interactions arise in gene regulatory networks has remained unclear; here the appearance of pheromone-response regulator binding sites in the regulatory DNA of the a-specific genes of Saccharomyces cerevisiae are shown to have required specific changes in a second pathway during the evolution from its common ancestor with Candida albicans.
- Trevor R. Sorrells
- , Lauren N. Booth
- & Alexander D. Johnson
-
Article
| Open Access Transcription factor binding dynamics during human ES cell differentiation
Transcription factor binding dynamics during human ES cell differentiation
Lineage-specific transcription factors and signalling pathways cooperate with pluripotency regulators to control the transcriptional networks that drive cell specification and exit from an embryonic stem cell state; here, we report genome-wide binding data for 38 transcription factors combined with analysis of epigenomic and gene expression data during the differentiation of human embryonic stem cells into the three germ layers.
- Alexander M. Tsankov
- , Hongcang Gu
- & Alexander Meissner
-
Article |
 Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
The CRISPR-Cas9 system, a powerful tool for genome editing, has been engineered to activate endogenous gene transcription specifically and potently on a genome-wide scale and applied to a large-scale gain-of-function screen for studying melanoma drug resistance.
- Silvana Konermann
- , Mark D. Brigham
- & Feng Zhang
-
Letter |
 R-loops induce repressive chromatin marks over mammalian gene terminators
R-loops induce repressive chromatin marks over mammalian gene terminators
R-loops, which have been considered to be rare and potentially harmful transcriptional by-products, are now shown to be needed for antisense transcription and to induce repressive chromatin marks that reinforce pausing of transcription and thereby enhance its termination.
- Konstantina Skourti-Stathaki
- , Kinga Kamieniarz-Gdula
- & Nicholas J. Proudfoot
-
Letter |
 Regulation of RNA polymerase II activation by histone acetylation in single living cells
Regulation of RNA polymerase II activation by histone acetylation in single living cells
The interplay of histone acetylation and RNA polymerase II activity is investigated using fluorescence microscopy; acetylation of H3 at Lys 27 enhances the recruitment of a transcriptional activator and accelerates the transition of RNA polymerase II from initiation to elongation, thus indicating that histone acetylation has a causal effect on two distinct steps in transcription activation.
- Timothy J. Stasevich
- , Yoko Hayashi-Takanaka
- & Hiroshi Kimura
-
Letter |
 Required enhancer–matrin-3 network interactions for a homeodomain transcription program
Required enhancer–matrin-3 network interactions for a homeodomain transcription program
The POU homeodomain transcription factor Pit1 is required for pituitary development; here Pit1-occupied enhancers are shown to interact with the nuclear architecture components matrin-3 and Satb1, and this association is required for activation of Pit1-regulated enhancers and coding target genes.
- Dorota Skowronska-Krawczyk
- , Qi Ma
- & Michael G. Rosenfeld
-
Brief Communications Arising |
 Universality of core promoter elements?
Universality of core promoter elements?
- Matthias Siebert
- & Johannes Söding
-
Letter |
 Enhancer loops appear stable during development and are associated with paused polymerase
Enhancer loops appear stable during development and are associated with paused polymerase
A high-resolution map of enhancer three-dimensional contacts during Drosophila embryogenesis shows that although local regulatory interactions are frequent, long-range interactions are also very common; unexpectedly, most interactions appear unchanged between tissues and across development and are formed prior to gene expression, indicating that transcription initiates from preformed enhancer–promoter loops, which are associated with paused polymerase.
- Yad Ghavi-Helm
- , Felix A. Klein
- & Eileen E. M. Furlong
-
Letter |
 Obesity-associated variants within FTO form long-range functional connections with IRX3
Obesity-associated variants within FTO form long-range functional connections with IRX3
Obesity-associated noncoding sequences within FTO are functionally connected with IRX3, and long-range enhancers in this region recapitulate aspects of IRX3 expression, suggesting that the obesity-associated interval is part of IRX3 regulation; Irx3-deficient mice have lower body weight and are resistant to diet-induced obesity, suggesting IRX3 as a novel determinant of body mass and composition.
- Scott Smemo
- , Juan J. Tena
- & Marcelo A. Nóbrega
-
Letter |
 Convergent evolution of a fused sexual cycle promotes the haploid lifestyle
Convergent evolution of a fused sexual cycle promotes the haploid lifestyle
In the predominantly diploid yeast Saccharomyces cerevisiae, regulatory control of mating is separate from meiosis; here the related hemiascomycete yeast Candida lusitaniae is shown to have coordinated regulatory control of mating and meiosis, favouring the formation of haploids.
- Racquel Kim Sherwood
- , Christine M. Scaduto
- & Richard J. Bennett

 Affinity-optimizing enhancer variants disrupt development
Affinity-optimizing enhancer variants disrupt development