Featured
-
-
Article |
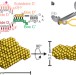 The structure of the box C/D enzyme reveals regulation of RNA methylation
The structure of the box C/D enzyme reveals regulation of RNA methylation
RNAs undergo many types of post-transcriptional modification, including methylation of ribosomal RNAs; here the structure of the archaeal box C/D ribonucleoprotein complex bound to substrate RNA is determined, showing that the two methylation guide sequences exist in different contexts and revealing sequential regulation of methylation at the two sites.
- Audrone Lapinaite
- , Bernd Simon
- & Teresa Carlomagno
-
Letter |
 Three-state mechanism couples ligand and temperature sensing in riboswitches
Three-state mechanism couples ligand and temperature sensing in riboswitches
In the human pathogen Vibrio vulnificus, the riboswitch regulating gene expression of the adenosine deaminase is shown to exist in three distinct stable conformational states; this three-state mechanism allows control of gene expression over a broad temperature range, which is essential for Vibrio adaptation.
- Anke Reining
- , Senada Nozinovic
- & Harald Schwalbe
-
Article |
 A compendium of RNA-binding motifs for decoding gene regulation
A compendium of RNA-binding motifs for decoding gene regulation
This study reports a global analysis of binding sites for over 200 RNA-binding proteins (RBPs) from 24 species; conserved RNA-binding motifs are identified, and their analysis allows prediction of interaction sites based on the sequence of the RNA-binding domain alone.
- Debashish Ray
- , Hilal Kazan
- & Timothy R. Hughes
-
Letter |
 Unusual base pairing during the decoding of a stop codon by the ribosome
Unusual base pairing during the decoding of a stop codon by the ribosome
Here, the structure of the 30S ribosomal subunit and the 70S ribosome in complex with a messenger RNA with pseudouridine in the place of uridine reveals unexpected base pairing.
- Israel S. Fernández
- , Chyan Leong Ng
- & V. Ramakrishnan
-
Letter |
 MBNL proteins repress ES-cell-specific alternative splicing and reprogramming
MBNL proteins repress ES-cell-specific alternative splicing and reprogramming
This study identifies MBNL proteins as negative regulators of alternative splicing events that are differentially regulated between ES cells and other cell types; several lines of evidence show that these proteins repress an ES cell alternative splicing program and the reprogramming of somatic cells to induced pluripotent stem cells.
- Hong Han
- , Manuel Irimia
- & Benjamin J. Blencowe
-
Letter |
 Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Members of the SAM-dependent methyltransferase superfamily are involved in the modification of wobble uridine to 5-oxacetyl uridine in Gram-negative bacteria; CmoA converts SAM to carboxy-SAM (Cx-SAM; a metabolite that was unknown previously), and CmoB uses Cx-SAM to convert 5-hydroxyuridine to 5-oxyacetyl uridine in tRNA.
- Jungwook Kim
- , Hui Xiao
- & Steven C. Almo
-
Letter |
 A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28–let-7 pathway
A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28–let-7 pathway
This study shows that Dis3l2 is the 3′–5′ exonuclease that mediates the degradation of uridylated precursor let-7 microRNA; this is the first physiological RNA substrate identified for this new exonuclease, which causes the Perlman syndrome of fetal overgrowth and Wilms’ tumour susceptibility when mutated.
- Hao-Ming Chang
- , Robinson Triboulet
- & Richard I. Gregory
-
Letter |
 CPEB1 coordinates alternative 3′-UTR formation with translational regulation
CPEB1 coordinates alternative 3′-UTR formation with translational regulation
CPEB1 is known to regulate cytoplasmic polyadenylation, and is now shown to have a second function in the nucleus; it associates with the cleavage and polyadenylation machinery, thereby promoting usage of an upstream poly(A) signal in many messenger RNAs, and affecting alternative splicing.
- Felice-Alessio Bava
- , Carolina Eliscovich
- & Raúl Méndez
-
Letter |
 Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence
Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence
To identify comprehensively factors involved in RNAi and microRNA-mediated gene expression regulation, this study performed a phylogenetic analysis of 86 eukaryotic species; the candidates this approach highlighted were subjected to Bayesian analysis with transcriptional and proteomic interaction data, identifying protein orthologues of already known RNAi silencing factors, as well as other hits involved in splicing, suggesting a connection between the two processes.
- Yuval Tabach
- , Allison C. Billi
- & Gary Ruvkun
-
Article |
 FMRP targets distinct mRNA sequence elements to regulate protein expression
FMRP targets distinct mRNA sequence elements to regulate protein expression
RNA-recognition elements are identified for the fragile-X-syndrome-associated RNA-binding protein FMRP, in addition to its target messenger RNAs; although many of FMRP gene targets discovered are involved in brain function and autism spectrum disorder, a proportion are also dysregulated in mouse ovaries, suggesting cross-regulation of signalling pathways in different tissues.
- Manuel Ascano
- , Neelanjan Mukherjee
- & Thomas Tuschl
-
Letter |
 Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p
Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p
Analysis of the yeast DEAD-box nucleic acid helicase Mss116p provides a structural model for how DEAD-box proteins recognize and unwind RNA duplexes.
- Anna L. Mallam
- , Mark Del Campo
- & Alan M. Lambowitz
-
Letter |
 Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi
Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi
Translocation of glycolytic enzymes to peroxisomes in fungi suggests broader metabolic role for this organelle.
- Johannes Freitag
- , Julia Ast
- & Michael Bölker
-
Article |
 Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq
Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq
N6-methyladenosine (m6A) is the most prevalent internal modification in messenger RNA; here the human and mouse m6A modification landscape is presented in a transcriptome-wide manner, providing insights into this epigenetic modification.
- Dan Dominissini
- , Sharon Moshitch-Moshkovitz
- & Gideon Rechavi
-
Letter |
 DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation
DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation
Characterization of the human interactome of chromatin-associated messenger ribonucleoprotein particles identifies DBC1 and a new protein (ZIRD) as subunits of a protein complex (DBIRD) that binds directly to RNAPII, regulates alternative splicing of exons embedded in (A + T)-rich DNA, and whose depletion results in region-specific decreases in transcript elongation.
- Pierre Close
- , Philip East
- & Jesper Q. Svejstrup
-
News |
 RNA editing study under intense scrutiny
RNA editing study under intense scrutiny
Debate highlights pitfalls in interpreting genomic data.
- Erika Check Hayden
-
Article |
 CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing
CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing
- Sanjeev Shukla
- , Ersen Kavak
- & Shalini Oberdoerffer
-
News & Views |
 A hidden ancestral legacy trumped
A hidden ancestral legacy trumped
A previously unsuspected genetic mechanism underlies a type of muscular dystrophy common in Japan. A therapeutic approach based on this finding and tested in mice has come up with encouraging results. See Letter p.127
- Masayuki Nakamori
- & Charles Thornton
-
Letter |
 Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy
Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy
- Mariko Taniguchi-Ikeda
- , Kazuhiro Kobayashi
- & Tatsushi Toda
-
Letter |
 Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats
Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats
- Elena O. Gracheva
- , Julio F. Cordero-Morales
- & David Julius
-
Article |
 Dicer recognizes the 5′ end of RNA for efficient and accurate processing
Dicer recognizes the 5′ end of RNA for efficient and accurate processing
- Jong-Eun Park
- , Inha Heo
- & V. Narry Kim
-
Letter |
 Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF
Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF
- Cameron D. Mackereth
- , Tobias Madl
- & Michael Sattler
-
Letter |
 Converting nonsense codons into sense codons by targeted pseudouridylation
Converting nonsense codons into sense codons by targeted pseudouridylation
- John Karijolich
- & Yi-Tao Yu
-
News |
 Evidence of altered RNA stirs debate
Evidence of altered RNA stirs debate
Sceptics question find that upends biology's 'central dogma'.
- Erika Check Hayden
-
Article |
 Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing
Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing
- Shravan Kumar Mishra
- , Tim Ammon
- & Stefan Jentsch
-
News |
 Cells may stray from 'central dogma'
Cells may stray from 'central dogma'
The ability to edit RNA to produce 'new' protein-coding sequences could be widespread in human cells.
- Erika Check Hayden
-
Letter |
 Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis
Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis
- Adelaine K. W. Leung
- , Kiyoshi Nagai
- & Jade Li
-
Article |
 CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III
CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III
CRISPR is a microbial RNA-based immune system protecting against viral and plasmid invasions. The CRISPR system is thought to rely on cleavage of a precursor RNA transcript by Cas endonucleases, but not all species possessing CRISPR-type immunity encode Cas proteins. This study now describes an alternative pathway in Streptococcus pyogenes that employs trans-encoded small RNA that directs the processing of precursor RNA into crRNAs through endogenous RNase III and the CRISPR-associated Csn1 protein.
- Elitza Deltcheva
- , Krzysztof Chylinski
- & Emmanuelle Charpentier
-
Letter |
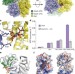 A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export
A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export
- Ben Montpetit
- , Nathan D. Thomsen
- & Karsten Weis
-
Letter |
 lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements
lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements
Staufen 1 (STAU1) protein binds regions of dsRNA in the 3′ UTR of mRNAs and promotes their degradation, a process known as SMD (Staufen-mediated mRNA decay). Although a specific stem-loop binding site had been defined for one SMD target, it was unclear how STAU1 was directed to other SMD targets that lack this structure. This paper reports that pairing of Alu element sequences in long non-coding RNAs (lncRNAs) and in the 3′ UTR of the SMD target generates a dsRNA structure that STAU1 recognizes. This result highlights a new function for lncRNAs.
- Chenguang Gong
- & Lynne E. Maquat
-
Research Highlights |
 Epigenetics: What makes a queen bee?
Epigenetics: What makes a queen bee?
-
News |
 DNA sequence may be lost in translation
DNA sequence may be lost in translation
Researchers tackle a mysterious genetic phenomenon
- Alla Katsnelson
-
Letter |
 A methyl transferase links the circadian clock to the regulation of alternative splicing
A methyl transferase links the circadian clock to the regulation of alternative splicing
Various biological processes are entrained by the day–night cycle to occur at a specific time of day. One way the circadian system exerts these effects is through post-transcriptional regulation. These authors show that a protein that transfers methyl groups onto several spliceosome subunits, PRMT5, is regulated by the light–dark cycle. Methylation of these subunits affects alternative splicing of some genes, thus making them subject to circadian control.
- Sabrina E. Sanchez
- , Ezequiel Petrillo
- & Marcelo J. Yanovsky
-
Article |
 Single-molecule analysis of Mss116-mediated group II intron folding
Single-molecule analysis of Mss116-mediated group II intron folding
DEAD-box helicases use ATP hydrolysis to unwind duplex RNA and facilitate RNA or RNA–protein remodelling. One such helicase is Mss116, which targets a particular group II intron in RNA. Here, single-molecule fluorescence was used to monitor the effect of Mss16 on a minimal construct containing this intron. The data show that Mss16 stimulates the sampling of different folded states of the RNA. Moreover, the helicase promotes RNA folding through discrete ATP-independent and ATP-dependent steps.
- Krishanthi S. Karunatilaka
- , Amanda Solem
- & David Rueda
-
Article |
 U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation
U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation
Splicing is carried out by a collection of protein–RNA complexes known as snRNPs. The spliceosome contains equal quantities of the U1, U2, U4, U5 and U6 snRNPs, but the U1 snRNP is made in levels excess to the amounts needed to form spliceosomes, leading to the idea that excess U1s might have splicing independent functions. Here it is shown that the U1 snRNA interacts with some pre mRNAs whose introns have cryptic polyadenylation sites. This interaction prevents premature termination and polyadenylation of the pre mRNA.
- Daisuke Kaida
- , Michael G. Berg
- & Gideon Dreyfuss
-
Letter |
 Identification of a quality-control mechanism for mRNA 5′-end capping
Identification of a quality-control mechanism for mRNA 5′-end capping
Following their synthesis, eukaryotic messenger RNAs have a 7-methylguanosine cap added to their 5′ ends to protect the mRNAs from degradation. Here it is shown that, in vitro and in yeast, caps lacking a methyl group are recognized by the Rai1 protein, which clips off the incomplete cap. The data provide evidence that Rai1 is part of a quality-control mechanism that monitors, and promotes the digestion of, aberrant mRNAs that might arise during stress conditions.
- Xinfu Jiao
- , Song Xiang
- & Megerditch Kiledjian
-
Article |
 Deciphering the splicing code
Deciphering the splicing code
The coding capacity of the genome is greatly expanded by the process of alternative splicing, which enables a single gene to produce more than one distinct protein. Can the expression of these different proteins be predicted from sequence data? Here, modelling based on information theory has been used to develop a 'splicing code', which can predict, with good accuracy, tissue-dependent changes in alternative splicing.
- Yoseph Barash
- , John A. Calarco
- & Brendan J. Frey
-
Letter |
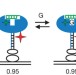 Multiple native states reveal persistent ruggedness of an RNA folding landscape
Multiple native states reveal persistent ruggedness of an RNA folding landscape
The 'thermodynamic hypothesis' proposes that the sequence of a biological macromolecule defines its folded, active structure as a global energy minimum in the folding landscape; however, it is not clear whether there is only one global minimum or several local minima corresponding to active conformations. Here, using single-molecule experiments, an RNA enzyme is shown to fold into multiple distinct native states that interconvert.
- Sergey V. Solomatin
- , Max Greenfeld
- & Daniel Herschlag
-
Review Article |
 Expansion of the eukaryotic proteome by alternative splicing
Expansion of the eukaryotic proteome by alternative splicing
- Timothy W. Nilsen
- & Brenton R. Graveley

 Crystal structure of the 14-subunit RNA polymerase I
Crystal structure of the 14-subunit RNA polymerase I