Article
|
Open Access
Featured
-
-
Article
| Open Access PLMSearch: Protein language model powers accurate and fast sequence search for remote homology
PLMSearch: Protein language model powers accurate and fast sequence search for remote homology
Homologous protein search is one of the most commonly used methods for protein analysis. Here, authors propose PLMSearch, a search method that takes only sequences as input and can search millions of protein pairs in seconds while maintaining sensitivity comparable to SOTA structure search methods.
- Wei Liu
- , Ziye Wang
- & Shanfeng Zhu
-
Article
| Open Access AlphaPept: a modern and open framework for MS-based proteomics
AlphaPept: a modern and open framework for MS-based proteomics
Mass spectrometry-based proteomics faces the challenge of processing vast data amounts. Here, the authors introduce AlphaPept, an open-source, Python-based framework that offers high speed analysis and easy integration for large-scale proteome analysis.
- Maximilian T. Strauss
- , Isabell Bludau
- & Matthias Mann
-
Article
| Open Access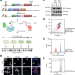 A mutational atlas for Parkin proteostasis
A mutational atlas for Parkin proteostasis
Gene variants can affect folding and stability of the encoded protein. Here, the authors apply deep mutational scanning to provide genotype-phenotype information for 99% of the possible PRKN variants and reveal mechanistic details on how some variants cause loss-of-function and Parkinsons disease.
- Lene Clausen
- , Vasileios Voutsinos
- & Rasmus Hartmann-Petersen
-
Article
| Open Access Local energetic frustration conservation in protein families and superfamilies
Local energetic frustration conservation in protein families and superfamilies
Energetic local frustration in proteins may have been positively selected by evolution when related to function such as ligand binding, allostery and other. Here the authors present a methodology to analyze local frustration patterns within protein families and superfamilies.
- Maria I. Freiberger
- , Victoria Ruiz-Serra
- & Alfonso Valencia
-
Article
| Open Access Defining the condensate landscape of fusion oncoproteins
Defining the condensate landscape of fusion oncoproteins
Many fusion oncoproteins (FOs) form condensates, some form in the nucleus and regulate gene expression while others form in the cytoplasm and promote cell signaling. In this work, the authors report the analysis of physicochemical features to enable prediction of FO condensation behavior.
- Swarnendu Tripathi
- , Hazheen K. Shirnekhi
- & Richard W. Kriwacki
-
Article
| Open Access Machine learning coarse-grained potentials of protein thermodynamics
Machine learning coarse-grained potentials of protein thermodynamics
Understanding protein dynamics is a complex scientific challenge. Here, authors construct coarse-grained molecular potentials using artificial neural networks, significantly accelerating protein dynamics simulations while preserving their thermodynamics.
- Maciej Majewski
- , Adrià Pérez
- & Gianni De Fabritiis
-
Article
| Open Access Global impact of somatic structural variation on the cancer proteome
Global impact of somatic structural variation on the cancer proteome
The relevance of non-coding somatic mutations in cancer remains elusive. Here, the combination of mass spectrometry-based proteomics and whole genome sequencing data across multiple cancer types helps to assess the effects of somatic structural variant breakpoint patterns on protein expression of nearby genes.
- Fengju Chen
- , Yiqun Zhang
- & Chad J. Creighton
-
Article
| Open Access Protein-Peptide Turnover Profiling reveals the order of PTM addition and removal during protein maturation
Protein-Peptide Turnover Profiling reveals the order of PTM addition and removal during protein maturation
Metabolic labeling is often used to measure protein turnover. Here the authors show that for interconvertible protein species like phosphoforms metabolic labeling does not provide information on turnover differences, but that the relative order of modification can determine the observed dynamics.
- Henrik M. Hammarén
- , Eva-Maria Geissen
- & Mikhail M. Savitski
-
Article
| Open Access Protein language models trained on multiple sequence alignments learn phylogenetic relationships
Protein language models trained on multiple sequence alignments learn phylogenetic relationships
Protein language models taking multiple sequence alignments as inputs capture protein structure and mutational effects. Here, the authors show that these models also encode phylogenetic relationships, and can disentangle correlations due to structural constraints from those due to phylogeny.
- Umberto Lupo
- , Damiano Sgarbossa
- & Anne-Florence Bitbol
-
Article
| Open Access Deciphering polymorphism in 61,157 Escherichia coli genomes via epistatic sequence landscapes
Deciphering polymorphism in 61,157 Escherichia coli genomes via epistatic sequence landscapes
Predicting the effects of mutations in a species is a major challenge in genetics. Here, the authors investigate protein sequence landscapes using diverged E. coli sequences, to predict tolerated mutations and capture interactions between mutations.
- Lucile Vigué
- , Giancarlo Croce
- & Martin Weigt
-
Article
| Open Access Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure
Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure
Most known pathogenic mutations occur in protein-coding regions of DNA and change the way proteins are made. Here the authors analyse the locations of thousands of human disease mutations and their predicted effects on protein structure and show that,while loss-of-function mutations tend to be highly disruptive, non-loss-of-function mutations are in general much milder at a protein structural level.
- Lukas Gerasimavicius
- , Benjamin J. Livesey
- & Joseph A. Marsh
-
Article
| Open Access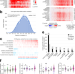 Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways
Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways
Pan-cancer proteomics analysis enables the analysis of protein expression across multiple cancer types. Here, the authors compare proteomics from 14 cancer types and show 11 distinct subtypes across multiple cancer types. Proteome data could link higher pathway activity levels with somatic alteration of specific genes in the pathway.
- Yiqun Zhang
- , Fengju Chen
- & Chad J. Creighton
-
Article
| Open Access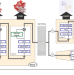 Mapping the glycosyltransferase fold landscape using interpretable deep learning
Mapping the glycosyltransferase fold landscape using interpretable deep learning
Glycosyltransferases (GT) are proteins that display extensive sequence and functional variation on a subset of 3D folds. Here, the authors use interpretable deep learning to predict 3D folds from sequence without the need for sequence alignment, which also enables the prediction of GTs with new folds.
- Rahil Taujale
- , Zhongliang Zhou
- & Natarajan Kannan
-
Article
| Open Access flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions
flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions
The authors present flDPnn, a computational tool for disorder and disorder function predictions from protein sequences. flDPnn was assessed with the data from the “Critical Assessment of Protein Intrinsic Disorder Prediction” experiment and on an independent and low-similarity test dataset, which show that flDPnn offers accurate predictions of disorder, fully disordered proteins and four common disorder functions.
- Gang Hu
- , Akila Katuwawala
- & Lukasz Kurgan
-
Article
| Open Access Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences
Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences
Our understanding of the residue-level details of protein interactions remains incomplete. Here, the authors show sequence coevolution can be used to infer interacting proteins with residue-level details, including predicting 467 interactions de novo in the Escherichia coli cell envelope proteome.
- Anna G. Green
- , Hadeer Elhabashy
- & Debora S. Marks
-
Article
| Open Access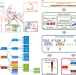 Inferring the molecular and phenotypic impact of amino acid variants with MutPred2
Inferring the molecular and phenotypic impact of amino acid variants with MutPred2
Identifying variants capable of causing genetic disease is challenging. The authors use semisupervised learning to predict pathogenic missense variants and their impacts on protein structure and function, enabling a molecular mechanism-driven approach to studying different types of human disease.
- Vikas Pejaver
- , Jorge Urresti
- & Predrag Radivojac
-
Article
| Open Access Molecular determinants underlying functional innovations of TBP and their impact on transcription initiation
Molecular determinants underlying functional innovations of TBP and their impact on transcription initiation
The TATA-box binding protein (TBP) is required for transcription initiation in archaea and eukaryotes. Here the authors delineate how TBP’s function has evolved new functional features through context-dependent interactions with various protein partners.
- Charles N. J. Ravarani
- , Tilman Flock
- & Santhanam Balaji
-
Article
| Open Access DIP/Dpr interactions and the evolutionary design of specificity in protein families
DIP/Dpr interactions and the evolutionary design of specificity in protein families
Dpr (Defective proboscis extension response) and DIP (Dpr Interacting Proteins) are immunoglobulin-like cell-cell adhesion proteins that form highly specific pairwise interactions, which control synaptic connectivity during Drosophila development. Here, the authors combine a computational approach with binding affinity measurements and find that DIP/Dpr binding specificity is controlled by negative constraints that interfere with non-cognate binding.
- Alina P. Sergeeva
- , Phinikoula S. Katsamba
- & Barry Honig
-
Article
| Open Access Dimensionality reduction by UMAP to visualize physical and genetic interactions
Dimensionality reduction by UMAP to visualize physical and genetic interactions
Dimensionality reduction is often used to visualize expression profiling data in order to find relationships among cells. Here, the authors use Uniform Manifold Approximation and Projection (UMAP) on published expression data of gene deletions of S. cerevisiae to find novel protein interactions.
- Michael W. Dorrity
- , Lauren M. Saunders
- & Cole Trapnell
-
Article
| Open Access Membrane protein-regulated networks across human cancers
Membrane protein-regulated networks across human cancers
Membrane proteins have been implicated in cancers, but studying the downstream effects of their perturbation remains challenging. Here, the authors map the membrane protein-regulated network of 15 cancers, a resource for prognostic biomarker development and druggable target identification.
- Chun-Yu Lin
- , Chia-Hwa Lee
- & Jinn-Moon Yang
-
Article
| Open Access A systematic approach to orient the human protein–protein interaction network
A systematic approach to orient the human protein–protein interaction network
The directions of most human protein-protein interactions (PPIs) remain unknown. Here, the authors use cancer genomic and drug response data to infer the direction of signal flow in the human PPI network and show that the directed network improves drug target and cancer driver gene prioritization.
- Dana Silverbush
- & Roded Sharan
-
Article
| Open Access Haplosaurus computes protein haplotypes for use in precision drug design
Haplosaurus computes protein haplotypes for use in precision drug design
Proteoforms arise as protein isoforms or as protein haplotypes, which are the result of genetic variation. Here, the authors develop Haplosaurus, a database that computes protein haplotypes genome-wide from existing genotype data and analyse protein haplotype variability in the 1000 Genomes dataset.
- William Spooner
- , William McLaren
- & Catherine Chaillan Huntington
-
Article
| Open Access Clustering huge protein sequence sets in linear time
Clustering huge protein sequence sets in linear time
Billions of metagenomic and genomic sequences fill up public datasets, which makes similarity clustering an important and time-critical analysis step. Here, the authors develop Linclust, an algorithm with linear time complexity that can cluster over a billion sequences within hours on a single server.
- Martin Steinegger
- & Johannes Söding
-
Article
| Open Access In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design
In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design
Antimicrobial peptides are considered promising alternatives to antibiotics. Here the authors developed a computational algorithm that starts with peptides naturally occurring in plants and optimizes this starting material to yield new variants which are highly distinct from the parent peptide.
- William F. Porto
- , Luz Irazazabal
- & Octavio L. Franco
-
Article
| Open Access CellCycleTRACER accounts for cell cycle and volume in mass cytometry data
CellCycleTRACER accounts for cell cycle and volume in mass cytometry data
Mass cytometry is a powerful method of single cell analysis, but potential confounding effects of cell cycle and cell volume are not taken into account. Here the authors present a combined experimental and computational method to correct for these effects and reveal features of TNFα stimulation that are otherwise masked.
- Maria Anna Rapsomaniki
- , Xiao-Kang Lun
- & María Rodríguez Martínez
-
Article
| Open Access De-novo protein function prediction using DNA binding and RNA binding proteins as a test case
De-novo protein function prediction using DNA binding and RNA binding proteins as a test case
Identification of the function of proteins is difficult when there are no structurally or biochemically characterized homologs. Here, the authors present an approach that allows the prediction of nucleic-acid binding proteins based on sequence alone, and they are able to experimentally validate their method.
- Sapir Peled
- , Olga Leiderman
- & Yanay Ofran
-
Article
| Open Access Protein analysis by time-resolved measurements with an electro-switchable DNA chip
Protein analysis by time-resolved measurements with an electro-switchable DNA chip
The comprehensive bioanalysis of proteins usually requires multi-step surface and mobile phase measurements. Here, the authors use chips functionalized with dynamically actuated nanolevers—DNA strands that can be switched in an electric field—to obtain motional dynamic measurements of proteins on a chip.
- Andreas Langer
- , Paul A. Hampel
- & Ulrich Rant
-
Article |
 Plasmonic substrates for multiplexed protein microarrays with femtomolar sensitivity and broad dynamic range
Plasmonic substrates for multiplexed protein microarrays with femtomolar sensitivity and broad dynamic range
Protein microarrays are useful both in basic research and also in disease monitoring and diagnosis, but their dynamic range is limited. By using plasmonic gold substrates with near-infrared fluorescent enhancement, Tabakman et al. demonstrate a multiplexed protein array with improved detection limits and dynamic range.
- Scott M. Tabakman
- , Lana Lau
- & Hongjie Dai

 The genetic landscape of a metabolic interaction
The genetic landscape of a metabolic interaction