Article
|
Open Access
Featured
-
-
Article
| Open Access Engineering self-deliverable ribonucleoproteins for genome editing in the brain
Engineering self-deliverable ribonucleoproteins for genome editing in the brain
The delivery of CRISPR RNPs has potential advantages over other genome editing approaches, including reduced off-target editing and reduced immunogenicity. Here the authors report self-deliverable Cas9 RNPs capable of robustly editing cultured cells in vitro and the mouse brain upon direct injections.
- Kai Chen
- , Elizabeth C. Stahl
- & Jennifer A. Doudna
-
Article
| Open Access Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Existing TadA-derived CBEs exhibit residual A•T-to-G•C editing activity and suffer from lower activity at several sequence contexts and with non-SpCas9 targeting domains. Here, the authors use phage-assisted evolution to evolve CBE6 variants that address these limitations.
- Emily Zhang
- , Monica E. Neugebauer
- & David R. Liu
-
Editorial
| Open AccessEfficient genetic improvement of orphan crops cannot follow the old path
Orphan crops hold the potential to diversify our food systems. Considering their unique characteristics, our deep understanding of major crops, and the availability of modern genomic tools, taking a different research path from what major crops have gone through could accelerate the genetic improvement of orphan crops.
-
Article
| Open Access CRISPR-based gene drives generate super-Mendelian inheritance in the disease vector Culex quinquefasciatus
CRISPR-based gene drives generate super-Mendelian inheritance in the disease vector Culex quinquefasciatus
Culex mosquitoes are carriers of major diseases like West Nile virus and are a public health concern. Here the authors present a CRISPR-Cas9 gene drive as a control technology in the Culex quinquefasciatus mosquito species.
- Tim Harvey-Samuel
- , Xuechun Feng
- & Valentino M. Gantz
-
Article
| Open Access Developing mitochondrial base editors with diverse context compatibility and high fidelity via saturated spacer library
Developing mitochondrial base editors with diverse context compatibility and high fidelity via saturated spacer library
Ddd-Aderived cytosine base editors (DdCBEs) are important for research of mitochondrial DNA mutation diseases. Here the authors report a strategy for screening and characterising dsDNA cytidine deaminases, and identify 7 DddA homologs which they optimise to minimise nuclear and mitochondrial off-target editing.
- Haifeng Sun
- , Zhaojun Wang
- & Bin Shen
-
Article
| Open Access Efficient plant genome engineering using a probiotic sourced CRISPR-Cas9 system
Efficient plant genome engineering using a probiotic sourced CRISPR-Cas9 system
In the field of plant genome engineering, new nucleases with improved editing efficiency and alterative PAM requirements are needed. Here, the authors report a probiotic sourced CRISPR-LrCas9 system with similar PAM requirement to Cas12a and show its high efficiencies in various genome editing applications.
- Zhaohui Zhong
- , Guanqing Liu
- & Yong Zhang
-
Article
| Open Access tgCRISPRi: efficient gene knock-down using truncated gRNAs and catalytically active Cas9
tgCRISPRi: efficient gene knock-down using truncated gRNAs and catalytically active Cas9
CRISPRi is used for gene silencing in mammalian cells. Here the authors report a gene-suppression/activation strategy using active Cas9 complexed with truncated gRNAs (tgCRISPRi/a) without causing DNA cleavage: they use this to repress or activate expression of several target genes throughout somatic tissues in Drosophila melanogaster.
- Ankush Auradkar
- , Annabel Guichard
- & Ethan Bier
-
Article
| Open Access A split and inducible adenine base editor for precise in vivo base editing
A split and inducible adenine base editor for precise in vivo base editing
TadA deaminases widely used in many base editors lack post-translational control in cells. Here the authors report a split adenine base editor (sABE) using chemically induced dimerisation (CID) to control the catalytic activity of TadA8e and show this can be used for PCSK9 gene editing in the mouse liver.
- Hongzhi Zeng
- , Qichen Yuan
- & Xue Gao
-
Article
| Open Access Programmable RNA detection with CRISPR-Cas12a
Programmable RNA detection with CRISPR-Cas12a
Cas12a is widely used in diagnostic platforms. Here the authors show that Cas12a can be programmed to directly detect RNA substrates, this is due to the 3’-end of the crRNA tolerating both RNA and DNA substrates: they use this to report a method, SAHARA, to detect RNA sequences.
- Santosh R. Rananaware
- , Emma K. Vesco
- & Piyush K. Jain
-
Article
| Open Access QTG-Miner aids rapid dissection of the genetic base of tassel branch number in maize
QTG-Miner aids rapid dissection of the genetic base of tassel branch number in maize
The lack of large-scale QTL cloning method hampers systematic dissection of genetic base of quantitative traits. Here, the authors develop a multi-omics data-based technique for large-scale and rapid cloning of quantitative genes of tassel branch number and discovery of selection signatures in maize breeding.
- Xi Wang
- , Juan Li
- & Lin Li
-
Article
| Open Access Whole genomic analysis reveals atypical non-homologous off-target large structural variants induced by CRISPR-Cas9-mediated genome editing
Whole genomic analysis reveals atypical non-homologous off-target large structural variants induced by CRISPR-Cas9-mediated genome editing
The safety of CRISPR-Cas9 editing is a concern. Here the authors use whole genomic analysis by 10x linked-read sequencing and optical genome mapping to interrogate the genome integrity after editing: they see large structural variants at on-target sites and unexpected large chromosomal deletions.
- Hsiu-Hui Tsai
- , Hsiao-Jung Kao
- & John Yu
-
Article
| Open Access A qPCR technology for direct quantification of methylation in untreated DNA
A qPCR technology for direct quantification of methylation in untreated DNA
Analysis of DNA methylation usually requires a chemical or an enzymatic pretreatment step. Here, the authors report a PCR-based technology for the detection of DNA methylation in untreated DNA, and present analytical and clinical results from methylation analysis of the MGMT promoter.
- Kamilla Kolding Bendixen
- , Maria Mindegaard
- & Rasmus Koefoed Petersen
-
Article
| Open Access SnapFISH: a computational pipeline to identify chromatin loops from multiplexed DNA FISH data
SnapFISH: a computational pipeline to identify chromatin loops from multiplexed DNA FISH data
Multiplexed DNA FISH technologies are powerful tools to reveal chromatin spatial organisation. Here, the authors developed SnapFISH, a computational pipeline to identify chromatin loops from multiplexed DNA FISH data.
- Lindsay Lee
- , Hongyu Yu
- & Ming Hu
-
Article
| Open Access HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing
HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing
Limited work has been done on concurrent C-to-G and A-to-G base editing. Here the authors test how a number of chromatin-associated factors affect base editing and show that HMGN1 enhanced the efficiency; by fusing HMGN1 to GBE and ABE they develop a CRISPR-based dual-function A-to-G and C-to-G base editor (GGBE).
- Chao Yang
- , Zhenzhen Ma
- & Xueli Zhang
-
Article
| Open Access Sequence terminus dependent PCR for site-specific mutation and modification detection
Sequence terminus dependent PCR for site-specific mutation and modification detection
Rapid and facile detection of specific nucleic acid modifications could have numerous applications. Here the authors present Specific Terminal Mediated Polymerase Chain Reaction (STEM-PCR) as a generic and accessible approach, and demonstrate proof-of-principle cancer biomarker detection.
- Gaolian Xu
- , Hao Yang
- & Hongchen Gu
-
Article
| Open Access A versatile, high-efficiency platform for CRISPR-based gene activation
A versatile, high-efficiency platform for CRISPR-based gene activation
The generation of CRISPR-mediated transcriptional activation (CRISPRa)-competent cell lines pose significant technical challenges. Here the authors report a platform for production of CRISPRa-ready cell populations which they combine with optimised expressed and synthetic gRNA scaffolds to enhance functionality.
- Amy J. Heidersbach
- , Kristel M. Dorighi
- & Benjamin Haley
-
Article
| Open Access DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing
DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing
There is a need to improve and expand mitochondrial base editing tools. Here the authors identify a DddA homolog from Simiaoa sunii (Ddd_Ss) which can efficiently deaminate cytosine in dsDNA; they develop cytosine base editors and introduce mutations at multiple mitochondrial DNA loci.
- Li Mi
- , Ming Shi
- & Yangming Wang
-
Article
| Open Access Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches
Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches
Application of CRISPR-Cas13d is limited by the inability to predict on- and off-targets. Here the authors perform CRISPR-Cas13d proliferation screens followed by modeling of Cas13d on- and off-targets; they design a deep learning model, DeepCas13, to predict the on-target activity of a gRNA.
- Xiaolong Cheng
- , Zexu Li
- & Wei Li
-
Article
| Open Access TadA orthologs enable both cytosine and adenine editing of base editors
TadA orthologs enable both cytosine and adenine editing of base editors
Properties of cytidine and adenosine deaminases lead to off-target effects for cytosine base editors (CBEs) and adenine base editors (ABEs). Here the authors report that 25 TadA orthologs could be engineered to generate functional ABEs, CBEs or ACBEs via single/double mutations with minimised off-targets.
- Shuqian Zhang
- , Bo Yuan
- & Tian-Lin Cheng
-
Article
| Open Access TadA reprogramming to generate potent miniature base editors with high precision
TadA reprogramming to generate potent miniature base editors with high precision
Hypercompact CRISPR-Cas12f systems have been engineered to generate miniABEs but these have limitations. Here the authors generate Cas12f-derived miniCBEs and develop miniABEs with improved editing and targeting scopes; they use these to correct pathogenic mutations in cell lines and introduce mutations in vivo.
- Shuqian Zhang
- , Liting Song
- & Tian-Lin Cheng
-
Article
| Open Access Development of a versatile nuclease prime editor with upgraded precision
Development of a versatile nuclease prime editor with upgraded precision
Strategies to improve the specificity of nuclease-based prime editor (PEn) are needed. Here the authors report a 53BP1-inhibitory ubiquitin variant-assisted PEn platform (uPEn) to inhibit NHEJ and enable precise prime editing for generation of insertions, deletions and replacements.
- Xiangyang Li
- , Guiquan Zhang
- & Xingxu Huang
-
Review Article
| Open Access Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing
Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing
CRISPR-Cas tools have shown exceptional promise in genome engineering over the past decade. Here the authors review the development of CRISPR-Cas9/Cas12/Cas13 nucleases, DNA base editors, prime editors, and RNA base editors, as well as their editing precision, off-target effects, and clinical considerations.
- Jianli Tao
- , Daniel E. Bauer
- & Roberto Chiarle
-
Article
| Open Access CRISPR-based targeted haplotype-resolved assembly of a megabase region
CRISPR-based targeted haplotype-resolved assembly of a megabase region
Low-cost targeted approach to construct haplotype-resolved assemblies is needed to facilitate population genetic studies. Here, the authors demonstrate assembling high-quality MHC haplotypes with CRISPR-based enrichment and long-read sequencings.
- Taotao Li
- , Duo Du
- & Yun Liu
-
Article
| Open Access TAPE-seq is a cell-based method for predicting genome-wide off-target effects of prime editor
TAPE-seq is a cell-based method for predicting genome-wide off-target effects of prime editor
Methods to predict genome-wide off-target activities of prime editors (PEs) are currently lacking. Here the authors report a cell-based assay, TAgmentation of Prime Editor sequencing (TAPE-seq), that provides genome-wide off-target candidates for PEs.
- Jeonghun Kwon
- , Minyoung Kim
- & Jungjoon K. Lee
-
Article
| Open Access Achieving single nucleotide sensitivity in direct hybridization genome imaging
Achieving single nucleotide sensitivity in direct hybridization genome imaging
Visualisation of point mutations in situ is informative for studying genetic diseases. Here the authors report single guide genome oligopaint via local denaturation fluorescence in situ hybridisation, sgGOLDFISH, a direct hybridisation genome imaging method with single-nucleotide sensitivity.
- Yanbo Wang
- , W. Taylor Cottle
- & Taekjip Ha
-
Article
| Open Access Automated high-throughput genome editing platform with an AI learning in situ prediction model
Automated high-throughput genome editing platform with an AI learning in situ prediction model
A large number of cell disease models with pathogenic SNVs are needed. Here the authors report an automated high-throughput platform to perform the genome editing process from gRNA design to the analysis of the editing results; they characterise in situ base editing outcomes.
- Siwei Li
- , Jingjing An
- & Meng Wang
-
Article
| Open Access Gene activation guided by nascent RNA-bound transcription factors
Gene activation guided by nascent RNA-bound transcription factors
Gene activation methods are valuable for studying gene functions and may have potential applications in bioengineering and medicine. Here the authors developed Narta technology to achieve gene activation by recruiting artificial transcription factors to transcription sites through nascent RNAs of the target gene.
- Ying Liang
- , Haiyue Xu
- & Baohui Chen
-
Article
| Open Access A p38α-BLIMP1 signalling pathway is essential for plasma cell differentiation
A p38α-BLIMP1 signalling pathway is essential for plasma cell differentiation
Plasma cells are terminally differentiated B cells that are specialized for antibody secretion. Authors show here that genomic deletion of the p38α mitogen activated protein kinase specifically in the B cell lineage leads to diminished plasma cell differentiation via impairment of a transcriptional regulatory program by BLIMP1.
- Jianfeng Wu
- , Kang Yang
- & Jiahuai Han
-
Article
| Open Access Identification of purine biosynthesis as an NADH-sensing pathway to mediate energy stress
Identification of purine biosynthesis as an NADH-sensing pathway to mediate energy stress
Reductive stress, reflected by the elevated intracellular NADH/NAD+ ratio, is associated with multiple human diseases. Here, the authors develop a genetic tool to manipulate the ratios of cellular NADH/NAD+ and NADPH/NADP+, and identify purine biosynthesis as an NADH-sensing pathway to mediate reductive stress.
- Ronghui Yang
- , Chuanzhen Yang
- & Binghui Li
-
Article
| Open Access SuperFi-Cas9 exhibits remarkable fidelity but severely reduced activity yet works effectively with ABE8e
SuperFi-Cas9 exhibits remarkable fidelity but severely reduced activity yet works effectively with ABE8e
Increased-fidelity SpCas9 variants have been developed, but often show proportionally reduced activity. Here the authors characterise the on-target activity and off-target propensity of SuperFi-Cas9 in mammalian cells and also see strongly reduced activity but with high fidelity features.
- Péter István Kulcsár
- , András Tálas
- & Ervin Welker
-
Article
| Open Access A comprehensive Bioconductor ecosystem for the design of CRISPR guide RNAs across nucleases and technologies
A comprehensive Bioconductor ecosystem for the design of CRISPR guide RNAs across nucleases and technologies
The success of CRISPR experiments relies on the choice of gRNA. Here the authors report crisprVerse, which enables efficient gRNA design and annotation for methods including CRISPRko, CRISPRa, CRISPRi, CRISPRbe and CRISPRkd, enabled for RNA- and DNA-targeting nucleases, including Cas9, Cas12 and Cas13.
- Luke Hoberecht
- , Pirunthan Perampalam
- & Jean-Philippe Fortin
-
Article
| Open Access Engineered helicase replaces thermocycler in DNA amplification while retaining desired PCR characteristics
Engineered helicase replaces thermocycler in DNA amplification while retaining desired PCR characteristics
PCR is an essential method for the amplification and manipulation of nucleic acids, but the requirement for a thermocycler limits access. Here, authors engineer a helicase to replace the heating step of PCR with enzymatic unwinding, allowing the isothermal amplification of fragments up to 6 kb.
- Momčilo Gavrilov
- , Joshua Y. C. Yang
- & Taekjip Ha
-
Article
| Open Access CtIP-dependent nascent RNA expression flanking DNA breaks guides the choice of DNA repair pathway
CtIP-dependent nascent RNA expression flanking DNA breaks guides the choice of DNA repair pathway
RNA has been implicated in DNA repair. This work shows that the interplay of RNAPII-generated nascent RNA, RNA:DNA hybrids and the resection factor CtIP guide DNA double strand break repair pathway choice towards error-free homologous recombination.
- Daniel Gómez-Cabello
- , George Pappas
- & Jiri Bartek
-
Article
| Open Access Accounting for small variations in the tracrRNA sequence improves sgRNA activity predictions for CRISPR screening
Accounting for small variations in the tracrRNA sequence improves sgRNA activity predictions for CRISPR screening
Existing methods for generating sgRNA predictions do not account for the tracrRNA sequence. Here the authors report an on-target model, Rule Set 3, to generate optimal predictions for multiple tracrRNA variants, and validate this on a new dataset of sgRNAs showing improvement over prior prediction models.
- Peter C. DeWeirdt
- , Abby V. McGee
- & John G. Doench
-
Article
| Open Access Recursive Editing improves homology-directed repair through retargeting of undesired outcomes
Recursive Editing improves homology-directed repair through retargeting of undesired outcomes
CRISPR-Cas induced HDR methods tend to have a low efficiency. Here the authors report an HDR improvement strategy, Recursive Editing, that selectively retargets undesired indel outcomes to create additional opportunities for HDR; they introduce REtarget, a tool for Recursive Editing experimental design.
- Lukas Möller
- , Eric J. Aird
- & Jacob E. Corn
-
Article
| Open Access Detection of neutralizing antibodies against multiple SARS-CoV-2 strains in dried blood spots using cell-free PCR
Detection of neutralizing antibodies against multiple SARS-CoV-2 strains in dried blood spots using cell-free PCR
Neutralizing antibodies are critical for conferring immunity against SARS-CoV-2. Here, Dahn et al. report a PCR assay termed SONIA (Split-Oligonucleotide Neighboring Inhibition Assay) for measuring neutralizing antibodies against multiple SARS-CoV-2 strains in fingerprick dried blood spot samples.
- Kenneth Danh
- , Donna Grace Karp
- & Cheng-ting Tsai
-
Article
| Open Access Efficient spatially targeted gene editing using a near-infrared activatable protein-conjugated nanoparticle for brain applications
Efficient spatially targeted gene editing using a near-infrared activatable protein-conjugated nanoparticle for brain applications
Spatial control of gene expression allows precise control over biological processes. Here, the authors develop an efficient light-responsive formulation based on upconversion nanoparticles, and demonstrate on-demand genetic manipulation in deep brain tissue.
- Catarina Rebelo
- , Tiago Reis
- & Lino Ferreira
-
Article
| Open Access Ultra-sensitive monitoring of leukemia patients using superRCA mutation detection assays
Ultra-sensitive monitoring of leukemia patients using superRCA mutation detection assays
Rare tumour specific mutations in patient samples act as markers to monitor the course of disease. Here the authors report superRCA assays for rapid, highly specific detection of DNA sequence variants present at very low frequencies in DNA samples with flow cytometry readout; they use this on AML patients.
- Lei Chen
- , Anna Eriksson
- & Ulf Landegren
-
Article
| Open Access PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction
PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction
Despite their broad utility, design and construction of plasmids remains laborious and time-consuming. Here the authors report a robust, versatile, and automated end-to-end platform that enables scarless construction of virtually any plasmid.
- Behnam Enghiad
- , Pu Xue
- & Huimin Zhao
-
Article
| Open Access Enhancement of prime editing via xrRNA motif-joined pegRNA
Enhancement of prime editing via xrRNA motif-joined pegRNA
The prime editors (PEs) have shown great promise for precise genome modification. Here the authors place a stabilizing viral xrRNA motif to the 3′ of pegRNAs to enhance editing efficiencies.
- Guiquan Zhang
- , Yao Liu
- & Jianghuai Liu
-
Article
| Open Access Phage peptides mediate precision base editing with focused targeting window
Phage peptides mediate precision base editing with focused targeting window
Base editors are genome engineering tools that can generate nucleotide substitutions without introducing double-stranded breaks. Here the authors show that a phage-derived peptidyl CRISPR inhibitor can be employed to modulate the activity and targeting scope of CRISPR base editor for precision base editing applications.
- Kun Jia
- , Yan-ru Cui
- & Jia Liu
-
Article
| Open Access Sonogenetic control of mammalian cells using exogenous Transient Receptor Potential A1 channels
Sonogenetic control of mammalian cells using exogenous Transient Receptor Potential A1 channels
Ultrasound can be used to non-invasively control neuronal functions. Here the authors report the use of human Transient receptor potential ankyrin 1 (hsTRPA1) to achieve ultrasound sensitivity in mammalian cells, and show that it can be used to manipulate neurons in the mammalian brain.
- Marc Duque
- , Corinne A. Lee-Kubli
- & Sreekanth H. Chalasani
-
Article
| Open Access Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9
Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9
The causal relationship between DNA demethylation and gene expression regulation has not yet been fully resolved. Here the authors develop a nuclease-dead Cas9 (dCas9) and gRNA site-specific targeting approach to physically block DNA methylation at specific promoters to cause DNA demethylation in cells and tackle this question.
- Daniel M. Sapozhnikov
- & Moshe Szyf
-
Article
| Open Access The identification of grain size genes by RapMap reveals directional selection during rice domestication
The identification of grain size genes by RapMap reveals directional selection during rice domestication
Cloning quantitative trait loci (QTL) in crops is often slow and laborious. Here, the authors describe RapMap, a method to rapidly clone multiple QTL based on F2 gradient populations coupled with a co-segregation standard, and show how it can be used to identify genes controlling grain size in rice.
- Juncheng Zhang
- , Dejian Zhang
- & Yibo Li
-
Article
| Open Access Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain
Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain
While prime editing is a promising technology, PE2 systems often have low efficiency. Here the authors fuse a Rad51 DNA-binding domain to create hyPE2 with improved editing efficiency.
- Myungjae Song
- , Jung Min Lim
- & Hyongbum Henry Kim
-
Article
| Open Access Small molecule inhibition of ATM kinase increases CRISPR-Cas9 1-bp insertion frequency
Small molecule inhibition of ATM kinase increases CRISPR-Cas9 1-bp insertion frequency
The mutational outcome of CRISPR-Cas9 editing can be both predictable and targeted. Here the authors show that ATM inhibitor KU-60019 increases 1 bp insertions at the targeted locus.
- Heysol C. Bermudez-Cabrera
- , Sannie Culbertson
- & Richard I. Sherwood
-
Article
| Open Access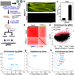 Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation
Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation
Throughput of in vivo genetic screens is a barrier to efficient application. Here the authors use a high-throughput CRISPR-based in vivo forward genetic screen in mice to identify transcriptional regulators of cardiomyocyte maturation, including the epigenetic modifiers RNF20 and RNF40.
- Nathan J. VanDusen
- , Julianna Y. Lee
- & William T. Pu
-
Article
| Open Access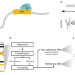 Cas9 targeted enrichment of mobile elements using nanopore sequencing
Cas9 targeted enrichment of mobile elements using nanopore sequencing
Mobile element insertions (MEIs) are a source of repetitive genetic variation and can lead to genetic disorders. Here the authors use Cas9-targeted nanopore sequencing to efficiently saturate enrichment for known and non-reference MEIs.
- Torrin L. McDonald
- , Weichen Zhou
- & Alan P. Boyle
-
Article
| Open Access Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning
Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning
High-quality gRNA activity data is needed for accurate on-target efficiency predictions. Here the authors generate activity data for over 10,000 gRNA and build a deep learning model CRISPRon for improved performance predictions.
- Xi Xiang
- , Giulia I. Corsi
- & Yonglun Luo

 dCas13-mediated translational repression for accurate gene silencing in mammalian cells
dCas13-mediated translational repression for accurate gene silencing in mammalian cells