Article
|
Open Access
Featured
-
-
Perspective |
 Hold out the genome: a roadmap to solving the cis-regulatory code
Hold out the genome: a roadmap to solving the cis-regulatory code
A roadmap towards solving the cis-regulatory code using a combination of machine learning and massively parallel assays of exogenous DNA is proposed.
- Carl G. de Boer
- & Jussi Taipale
-
Article
| Open Access Dissecting human population variation in single-cell responses to SARS-CoV-2
Dissecting human population variation in single-cell responses to SARS-CoV-2
Population differences in immune responses to SARS-CoV-2 can be explained by environmental exposures, but also by local adaptation acting through genetic variants acquired after admixture with archaic hominin forms.
- Yann Aquino
- , Aurélie Bisiaux
- & Lluis Quintana-Murci
-
Article |
 A spatially resolved single-cell genomic atlas of the adult human breast
A spatially resolved single-cell genomic atlas of the adult human breast
The Human Breast Cell Atlas identifies 12 major breast cell types and 58 biological cell states, revealing abundant pericyte, endothelial and immune cell populations, and highly diverse luminal epithelial cell states.
- Tapsi Kumar
- , Kevin Nee
- & Nicholas Navin
-
Article |
 Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours
Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours
A study identifies 41 consensus gene expression meta-programs that are coordinately upregulated in subpopulations of malignant cells across tumour types, providing a comprehensive picture of hallmarks of intratumour heterogeneity.
- Avishai Gavish
- , Michael Tyler
- & Itay Tirosh
-
Article
| Open Access Ageing-associated changes in transcriptional elongation influence longevity
Ageing-associated changes in transcriptional elongation influence longevity
Increases in transcriptional elongation speed with age affect organismal lifespan and ageing-related changes could be reversed with lifespan-extending interventions.
- Cédric Debès
- , Antonios Papadakis
- & Andreas Beyer
-
Article |
 Activation of γ-globin expression by hypoxia-inducible factor 1α
Activation of γ-globin expression by hypoxia-inducible factor 1α
Detailed mechanistic insight into fetal globin gene induction during hypoxia-associated stress erythropoiesis provides new therapeutic approaches to treat β-haemoglobinopathies, such as sickle cell disease and β-thalassaemia.
- Ruopeng Feng
- , Thiyagaraj Mayuranathan
- & Mitchell J. Weiss
-
Article
| Open Access Inferring and perturbing cell fate regulomes in human brain organoids
Inferring and perturbing cell fate regulomes in human brain organoids
A multi-omic atlas of brain organoid development facilitates the inference of an underlying gene regulatory network using the newly developed Pando framework and shows—in conjunction with perturbation experiments—that GLI3 controls forebrain fate establishment through interaction with HES4/5 regulomes.
- Jonas Simon Fleck
- , Sophie Martina Johanna Jansen
- & Barbara Treutlein
-
Article |
 Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy
Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy
- Mark Chaffin
- , Irinna Papangeli
- & Patrick T. Ellinor
-
Article |
 Synonymous mutations in representative yeast genes are mostly strongly non-neutral
Synonymous mutations in representative yeast genes are mostly strongly non-neutral
A survey of 8,341 mutations in 21 yeast genes shows that synonymous mutations are nearly as harmful as nonsynonymous mutations, in part because they both affect the mRNA level of the gene mutated.
- Xukang Shen
- , Siliang Song
- & Jianzhi Zhang
-
Article |
 Molecular hallmarks of heterochronic parabiosis at single-cell resolution
Molecular hallmarks of heterochronic parabiosis at single-cell resolution
A transcriptomics study demonstrates cell-type-specific responses to differentially aged blood and shows young blood to have restorative and rejuvenating effects that may be invoked through enhanced mitochondrial function.
- Róbert Pálovics
- , Andreas Keller
- & Tony Wyss-Coray
-
Article
| Open Access RNA profiles reveal signatures of future health and disease in pregnancy
RNA profiles reveal signatures of future health and disease in pregnancy
Expression signatures from cell-free RNA of pregnant women can be used to reveal normal biology of pregnancy and predict development of pre-eclampsia.
- Morten Rasmussen
- , Mitsu Reddy
- & Thomas F. McElrath
-
Article |
 Cardiopharyngeal deconstruction and ancestral tunicate sessility
Cardiopharyngeal deconstruction and ancestral tunicate sessility
The heart of appendicularians has evolved by 'deconstructing' an ancestral ascidian-like gene regulatory network.
- Alfonso Ferrández-Roldán
- , Marc Fabregà-Torrus
- & Cristian Cañestro
-
Matters Arising |
Phantom epistasis between unlinked loci
- Gibran Hemani
- , Joseph E. Powell
- & Peter M. Visscher
-
Review Article |
 Exploring tissue architecture using spatial transcriptomics
Exploring tissue architecture using spatial transcriptomics
This review describes the state of spatial transcriptomics technologies and analysis tools that are being used to generate biological insights in diverse areas of biology.
- Anjali Rao
- , Dalia Barkley
- & Itai Yanai
-
Article |
 Genome-wide enhancer maps link risk variants to disease genes
Genome-wide enhancer maps link risk variants to disease genes
Mapping enhancer regulation across human cell types and tissues illuminates genome function and provides a resource to connect risk variants for common diseases to their molecular and cellular functions.
- Joseph Nasser
- , Drew T. Bergman
- & Jesse M. Engreitz
-
Article |
 Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator
Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator
The long non-coding RNA locus Maenli controls mouse limb development by regulating En1 activity, and the absence of the homolgous MAENLI locus is associated with severe congenital limb defects in humans.
- Lila Allou
- , Sara Balzano
- & Andrea Superti-Furga
-
Article |
 Systematic analysis of binding of transcription factors to noncoding variants
Systematic analysis of binding of transcription factors to noncoding variants
An ultra-high-throughput multiplex protein–DNA binding assay is used to assess binding of 270 human transcription factors to 95,886 noncoding variants in the human genome, providing data to improve prediction of the effects of noncoding variants on transcription factor binding and thereby increase understanding of molecular pathways involved in diverse human traits and genetic diseases.
- Jian Yan
- , Yunjiang Qiu
- & Bing Ren
-
Article |
 A molecular cell atlas of the human lung from single-cell RNA sequencing
A molecular cell atlas of the human lung from single-cell RNA sequencing
Expression profiling on 75,000 single cells creates a comprehensive cell atlas of the human lung that includes 41 out of 45 previously known cell types and 14 new ones.
- Kyle J. Travaglini
- , Ahmad N. Nabhan
- & Mark A. Krasnow
-
Article |
 Transcriptome and translatome co-evolution in mammals
Transcriptome and translatome co-evolution in mammals
An analysis using ribosome-profiling and matched RNA-sequencing data for three organs across five mammalian species and a bird enables the comparison of translatomes and transcriptomes, revealing patterns of co-evolution of these two expression layers.
- Zhong-Yi Wang
- , Evgeny Leushkin
- & Henrik Kaessmann
-
Article
| Open Access Cells of the adult human heart
Cells of the adult human heart
Single-cell and single-nucleus RNA sequencing are used to construct a cellular atlas of the human heart that will aid further research into cardiac physiology and disease.
- Monika Litviňuková
- , Carlos Talavera-López
- & Sarah A. Teichmann
-
Article |
 Initiation of a conserved trophectoderm program in human, cow and mouse embryos
Initiation of a conserved trophectoderm program in human, cow and mouse embryos
Comparative analysis of human, cow and mouse embryos shows that a mechanism involving atypical protein kinase C initiates the trophectoderm program during the morula stage in these three species.
- Claudia Gerri
- , Afshan McCarthy
- & Kathy K. Niakan
-
Article |
 Histone H3.3 phosphorylation amplifies stimulation-induced transcription
Histone H3.3 phosphorylation amplifies stimulation-induced transcription
The histone variant H3.3 is phosphorylated at Ser31 in induced genes, and this selective mark stimulates the histone methyltransferase SETD2 and ejects the ZMYND11 repressor, thus revealing a role for histone phosphorylation in amplifying de novo transcription.
- Anja Armache
- , Shuang Yang
- & Steven Z. Josefowicz
-
Article |
 Ageing hallmarks exhibit organ-specific temporal signatures
Ageing hallmarks exhibit organ-specific temporal signatures
Bulk RNA sequencing of organs and plasma proteomics at different ages across the mouse lifespan is integrated with data from the Tabula Muris Senis, a transcriptomic atlas of ageing mouse tissues, to describe organ-specific changes in gene expression during ageing.
- Nicholas Schaum
- , Benoit Lehallier
- & Tony Wyss-Coray
-
Article |
 Mouse models of neutropenia reveal progenitor-stage-specific defects
Mouse models of neutropenia reveal progenitor-stage-specific defects
Mouse models of severe congenital neutropenia using patient-derived mutations in the GFI1 locus are used to determine the mechanisms by which the disease progresses.
- David E. Muench
- , Andre Olsson
- & H. Leighton Grimes
-
Letter |
 Reconstituting the transcriptome and DNA methylome landscapes of human implantation
Reconstituting the transcriptome and DNA methylome landscapes of human implantation
Transcriptomics and DNA methylomics are used to study the implantation process of human embryos at single-cell resolution.
- Fan Zhou
- , Rui Wang
- & Fuchou Tang
-
Letter |
 A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene
A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene
When Lama1 was upregulated using CRISPR and a catalytically inactive Cas9 in a mouse model of congenital muscular dystrophy type 1A, apparent hindlimb paralysis, muscle fibrosis and nerve myelination defects were ameliorated in symptomatic mice.
- Dwi U. Kemaladewi
- , Prabhpreet S. Bassi
- & Ronald D. Cohn
-
Letter |
 FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes
FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes
Mutations in the transcription factor FOXA1 that are common in prostate cancer result in gain-of-function effects that promote changes in the differentiation of tumour cells.
- Elizabeth J. Adams
- , Wouter R. Karthaus
- & Charles L. Sawyers
-
Review Article |
 Transcription factors and 3D genome conformation in cell-fate decisions
Transcription factors and 3D genome conformation in cell-fate decisions
Three-dimensional genome architecture has important roles in the regulation of gene expression and is therefore a key determinant of cell identity in normal development and in disease states.
- Ralph Stadhouders
- , Guillaume J. Filion
- & Thomas Graf
-
Letter |
 CDK12 regulates DNA repair genes by suppressing intronic polyadenylation
CDK12 regulates DNA repair genes by suppressing intronic polyadenylation
The kinase CDK12 suppresses usage of intronic polyadenylation sites and thereby promotes the expression of genes involved in homologous recombination DNA repair.
- Sara J. Dubbury
- , Paul L. Boutz
- & Phillip A. Sharp
-
Article |
 Shared and distinct transcriptomic cell types across neocortical areas
Shared and distinct transcriptomic cell types across neocortical areas
Single-cell transcriptomics of more than 20,000 cells from two functionally distinct areas of the mouse neocortex identifies 133 transcriptomic types, and provides a foundation for understanding the diversity of cortical cell types.
- Bosiljka Tasic
- , Zizhen Yao
- & Hongkui Zeng
-
Letter |
 Lineage tracking reveals dynamic relationships of T cells in colorectal cancer
Lineage tracking reveals dynamic relationships of T cells in colorectal cancer
An integrated RNA-sequencing approach demonstrates that CXCL13+ TH1-like cells are preferentially enriched in microsatellite-instable tumours from patients with colorectal cancer, and IGFLR1 is identified as a co-stimulatory molecule.
- Lei Zhang
- , Xin Yu
- & Zemin Zhang
-
Letter |
 A stromal cell population that inhibits adipogenesis in mammalian fat depots
A stromal cell population that inhibits adipogenesis in mammalian fat depots
Single-cell transcriptomics reveals that, in mice and humans, a population of cells in the stromal vascular fraction of adipose tissue regulates adipogenesis by suppressing adipocyte formation in a paracrine manner.
- Petra C. Schwalie
- , Hua Dong
- & Bart Deplancke
-
Article |
 Genomic atlas of the human plasma proteome
Genomic atlas of the human plasma proteome
A genetic atlas of the human plasma proteome, comprising 1,927 genetic associations with 1,478 proteins, identifies causes of disease and potential drug targets.
- Benjamin B. Sun
- , Joseph C. Maranville
- & Adam S. Butterworth
-
Letter |
 RNA polymerase III limits longevity downstream of TORC1
RNA polymerase III limits longevity downstream of TORC1
RNA polymerase III is a key evolutionarily conserved regulator of longevity that may have potential as a therapeutic target for age-related conditions.
- Danny Filer
- , Maximillian A. Thompson
- & Nazif Alic
-
Letter
| Open Access The impact of rare variation on gene expression across tissues
The impact of rare variation on gene expression across tissues
The authors show that rare genetic variants contribute to large gene expression changes across diverse human tissues and provide an integrative method for interpretation of rare variants in individual genomes.
- Xin Li
- , Yungil Kim
- & Stephen B. Montgomery
-
Article |
 Common genetic variation drives molecular heterogeneity in human iPSCs
Common genetic variation drives molecular heterogeneity in human iPSCs
Genetic and phenotypic analysis reveals expression quantitative trait loci in human induced pluripotent stem cell lines associated with cancer and disease.
- Helena Kilpinen
- , Angela Goncalves
- & Daniel J. Gaffney
-
Letter |
 Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism
Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism
Gene expression analysis in brain tissue from individuals with and without autism spectrum disorder (ASD) suggests that the transcription factor SOX5 contributes to an ASD-associated reduction in transcriptional differences between brain areas and indicates that common transcriptomic changes occur in different forms of ASD.
- Neelroop N. Parikshak
- , Vivek Swarup
- & Daniel H. Geschwind
-
Letter |
 Local regulation of gene expression by lncRNA promoters, transcription and splicing
Local regulation of gene expression by lncRNA promoters, transcription and splicing
Various cis-regulatory functions of genomic loci that produce long non-coding RNAs are revealed, including instances where their promoters have enhancer-like activity and the lncRNA transcripts themselves are not required for activity.
- Jesse M. Engreitz
- , Jenna E. Haines
- & Eric S. Lander
-
Letter |
 CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis
CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis
Atherosclerotic lesions in mice and humans switch on a ‘don’t eat me’ signal—expression of CD47—that prevents effective removal of diseased tissue; anti-CD47 antibody therapy can normalize this defective efferocytosis, with beneficial results in several mouse models of atherosclerosis.
- Yoko Kojima
- , Jens-Peter Volkmer
- & Nicholas J. Leeper
-
Letter |
 Resolving early mesoderm diversification through single-cell expression profiling
Resolving early mesoderm diversification through single-cell expression profiling
Analysis of the transcriptome of more than 1,200 cells from gastrulating mouse embryos using single-cell sequencing, gathering unexpected insights into early mesoderm formation during gastrulation.
- Antonio Scialdone
- , Yosuke Tanaka
- & Berthold Göttgens
-
Letter |
 Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease
Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease
Preclinical evaluation and optimization of mitochondrial replacement therapy reveals that a modified form of pronuclear transfer is likely to give rise to normal pregnancies with a reduced risk of mitochondrial DNA disease, but may need further modification to eradicate the disease in all cases.
- Louise A. Hyslop
- , Paul Blakeley
- & Mary Herbert
-
Letter |
 Translation readthrough mitigation
Translation readthrough mitigation
Translation termination sequences are occasionally bypassed by the ribosome and the resulting proteins can be detrimental to the cell; here it is shown that cells can prevent such proteins from accumulating through peptides that are encoded within the 3' UTR of genes in both humans and C. elegans.
- Joshua A. Arribere
- , Elif S. Cenik
- & Andrew Z. Fire
-
Letter |
 The industrial melanism mutation in British peppered moths is a transposable element
The industrial melanism mutation in British peppered moths is a transposable element
The mutation responsible for the black carbonaria morph of the peppered moth is identified as a transposable element within the cortex gene.
- Arjen E. van’t Hof
- , Pascal Campagne
- & Ilik J. Saccheri
-
Letter |
 Polygenic evolution of a sugar specialization trade-off in yeast
Polygenic evolution of a sugar specialization trade-off in yeast
An evolutionary trade-off of unprecedented genetic complexity in the glucose/galactose utilization regulatory pathway across several long-diverged species of Saccharomyces.
- Jeremy I. Roop
- , Kyu Chul Chang
- & Rachel B. Brem
-
Letter |
 The spliceosome is a therapeutic vulnerability in MYC-driven cancer
The spliceosome is a therapeutic vulnerability in MYC-driven cancer
Splicing factors such as BUD31 are identified in a synthetic-lethal screen with cells overexpressing the transcription factor MYC; oncogenic MYC leads to an increase in pre-mRNA synthesis, and spliceosome inhibition impairs the growth and tumorigenicity of MYC-dependent breast cancers, suggesting that spliceosome components may be potential therapeutic targets for MYC-driven cancers.
- Tiffany Y.-T. Hsu
- , Lukas M. Simon
- & Thomas F. Westbrook
-
Letter |
 Selection on noise constrains variation in a eukaryotic promoter
Selection on noise constrains variation in a eukaryotic promoter
Quantifying activity of cis-regulatory sequences controlling gene expression shows that selection on expression noise has a greater impact on sequence variation than selection on mean expression level.
- Brian P. H. Metzger
- , David C. Yuan
- & Patricia J. Wittkopp
-
Letter
| Open Access Integrative analysis of haplotype-resolved epigenomes across human tissues
Integrative analysis of haplotype-resolved epigenomes across human tissues
As part of the Epigenome Roadmap project, this study uses a chromosome-spanning haplotype reconstruction strategy to construct haplotype-resolved epigenomic maps for a diverse set of human tissues; the maps reveal extensive allelic biases in chromatin state and transcription, which vary across individuals due to genetic backgrounds.
- Danny Leung
- , Inkyung Jung
- & Bing Ren
-
Letter |
 Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation
Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation
The core promoters of developmental and housekeeping genes are shown to have distinct specificities for different enhancer sequences in Drosophila, and this specificity separates developmental and housekeeping gene regulatory programs across the genome.
- Muhammad A. Zabidi
- , Cosmas D. Arnold
- & Alexander Stark
-
Letter |
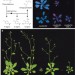 Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant
Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant
Disruption of lignin biosynthesis has been proposed as a way to improve forage and bioenergy crops, but it can result in stunted growth and developmental abnormalities; here, the undesirable features of one such manipulation are shown to depend on the transcriptional co-regulatory complex Mediator.
- Nicholas D. Bonawitz
- , Jeong Im Kim
- & Clint Chapple

 A single-cell time-lapse of mouse prenatal development from gastrula to birth
A single-cell time-lapse of mouse prenatal development from gastrula to birth