Featured
-
-
Article
| Open Access Impaired eIF5A function causes a Mendelian disorder that is partially rescued in model systems by spermidine
Impaired eIF5A function causes a Mendelian disorder that is partially rescued in model systems by spermidine
eIF5A is critical for protein synthesis but has not yet been associated with congenital human disease. Here, the authors show that EIF5A variants cause a Mendelian disorder via reduced eIF5A-ribosome interactions and this phenotype is partially corrected by spermidine supplementation in yeast and zebrafish.
- Víctor Faundes
- , Martin D. Jennings
- & Siddharth Banka
-
Article
| Open Access A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits
A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits
Statistical colocalisation is a method to identify causal genes and shared genetic aetiology across traits. Here, the authors describe HyPrColoc, an efficient Bayesian divisive clustering algorithm which integrates summary statistics from genome-wide association studies to detect clusters of colocalised traits from large numbers of traits.
- Christopher N. Foley
- , James R. Staley
- & Joanna M. M. Howson
-
Article
| Open Access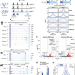 Conserved regulatory logic at accessible and inaccessible chromatin during the acute inflammatory response in mammals
Conserved regulatory logic at accessible and inaccessible chromatin during the acute inflammatory response in mammals
Genetic elements that control inflammatory gene expression are not fully elucidated. Here the authors conduct a multi-species analysis of chromatin landscape and NF-κB binding in response to the proinflammatory cytokine TNFα, finding that conserved NF-κB bound regions are linked to enhancer activity and disease.
- Azad Alizada
- , Nadiya Khyzha
- & Michael D. Wilson
-
Article
| Open Access White pupae phenotype of tephritids is caused by parallel mutations of a MFS transporter
White pupae phenotype of tephritids is caused by parallel mutations of a MFS transporter
The white pupae (wp) phenotype has been used for decades to selectively remove females of tephritid species in genetic sexing, but the determining gene is unknown. Here, the authors show that wp phenotype is produced by parallel mutations in a Major Facilitator Superfamily domain containing gene across multiple species.
- Christopher M. Ward
- , Roswitha A. Aumann
- & Marc F. Schetelig
-
Article
| Open Access Two novel venom proteins underlie divergent parasitic strategies between a generalist and a specialist parasite
Two novel venom proteins underlie divergent parasitic strategies between a generalist and a specialist parasite
Parasitism is a widespread evolutionary strategy. A study that spans functional and evolutionary genomics identifies the molecular basis and history underlying two genes that have mediated divergent parasitic strategies (specialist vs generalist) between two sister species of parasitoid wasp.
- Jianhua Huang
- , Jiani Chen
- & Shuai Zhan
-
Comment
| Open Access Functional studies of GWAS variants are gaining momentum
Functional studies of GWAS variants are gaining momentum
Rapidly advancing genomic technologies and cross-disciplinary partnerships are accelerating the biological and clinical interpretation of genome-wide association studies, with some therapies developed based on these findings already being tested in clinical trials. The next decade promises further progress in understanding the function of genetic variants.
- Florence Lichou
- & Gosia Trynka
-
Article
| Open Access Improving the informativeness of Mendelian disease-derived pathogenicity scores for common disease
Improving the informativeness of Mendelian disease-derived pathogenicity scores for common disease
Pathogenicity scores are instrumental in prioritizing variants for Mendelian disease, yet their application to common disease is largely unexplored. Here, the authors assess the utility of pathogenicity scores for 41 complex traits and develop a framework to improve their informativeness for common disease.
- Samuel S. Kim
- , Kushal K. Dey
- & Alkes L. Price
-
Article
| Open Access Construction of a complete set of Neisseria meningitidis mutants and its use for the phenotypic profiling of this human pathogen
Construction of a complete set of Neisseria meningitidis mutants and its use for the phenotypic profiling of this human pathogen
The bacterium Neisseria meningitidis causes life-threatening meningitis and sepsis. Here, Muir et al. construct a complete collection of defined mutants in protein-coding genes of this organism, which they use to identify its essential genome and to shed light on the functions of multiple genes.
- Alastair Muir
- , Ishwori Gurung
- & Vladimir Pelicic
-
Article
| Open Access Nuclear gene proximity and protein interactions shape transcript covariations in mammalian single cells
Nuclear gene proximity and protein interactions shape transcript covariations in mammalian single cells
Gene expression covariation can be studied by single-cell RNA sequencing. Here the authors analyze intrinsically covarying gene pairs by eliminating the confounding effects in single-cell experiments and observe covariation of proximal genes and miRNA-induced covariation of target mRNAs.
- Marcel Tarbier
- , Sebastian D. Mackowiak
- & Marc R. Friedländer
-
Article
| Open Access The structural variation landscape in 492 Atlantic salmon genomes
The structural variation landscape in 492 Atlantic salmon genomes
This study presents and validates a novel approach to reliably identify structural variations (SVs) in non-model genomes using whole genome sequencing, which was used to detect 15,483 SVs in 492 Atlantic salmon, shedding light on their roles in genome evolution and the genetic architecture of domestication.
- Alicia C. Bertolotti
- , Ryan M. Layer
- & Daniel J. Macqueen
-
Article
| Open Access Evaluating the informativeness of deep learning annotations for human complex diseases
Evaluating the informativeness of deep learning annotations for human complex diseases
Deep learning models have shown great promise in predicting regulatory effects from DNA sequence. Here the authors evaluate sequence-based epigenomic deep learning models and conclude that these models are not yet ready to inform our knowledge of human disease.
- Kushal K. Dey
- , Bryce van de Geijn
- & Alkes L. Price
-
Article
| Open Access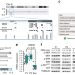 Haploinsufficiency of RREB1 causes a Noonan-like RASopathy via epigenetic reprogramming of RAS-MAPK pathway genes
Haploinsufficiency of RREB1 causes a Noonan-like RASopathy via epigenetic reprogramming of RAS-MAPK pathway genes
Mutations in RAS-MAPK pathway genes are implicated in Noonan-spectrum, yet up to 20% of cases have unknown cause. Here, the authors identify RREB1 underlying a 6p microdeletion RASopathy-like syndrome and show that RREB1, SIN3A and KDM1A form a transcriptional repressive complex to control methylation of MAPK pathway genes.
- Oliver A. Kent
- , Manipa Saha
- & Robert Rottapel
-
Article
| Open Access Analysis of chromatin organization and gene expression in T cells identifies functional genes for rheumatoid arthritis
Analysis of chromatin organization and gene expression in T cells identifies functional genes for rheumatoid arthritis
Although genome-wide association studies have identified genetic variation contributing to disease risk, assigning causal genes is challenging. Here, the authors generate ATAC-seq, Hi-C, Capture Hi-C and RNA-seq data in stimulated CD4+ T cells to identify functional enhancers and demonstrate interactions of expression quantitative trait loci with target genes in rheumatoid arthritis.
- Jing Yang
- , Amanda McGovern
- & Stephen Eyre
-
Article
| Open Access The genomic landscape of Mongolian hepatocellular carcinoma
The genomic landscape of Mongolian hepatocellular carcinoma
Mongolia has the highest incidence of—and mortality from—hepatocellular carcinoma (HCC) in the world. Here, the authors examine the genomic and transcriptomic landscape of Mongolian HCC, uncover novel driver mutations, and suggest distinct disease etiologies.
- Julián Candia
- , Enkhjargal Bayarsaikhan
- & Xin Wei Wang
-
Article
| Open Access Defining the ATPome reveals cross-optimization of metabolic pathways
Defining the ATPome reveals cross-optimization of metabolic pathways
Energy metabolism and ATP levels are controlled by an interlocking network of pathways. Here, the authors apply a genome-wide CRISPR screen to define genes that increase or decrease ATP levels to define the “ATPome”, a map of pathways that contribute to cellular ATP regulation.
- Neal K. Bennett
- , Mai K. Nguyen
- & Ken Nakamura
-
Article
| Open Access Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action
Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action
Large-scale screens of chemical and genetic vulnerabilities in cancer are typically limited to simple readouts of cell viability. Here, the authors develop a method for profiling post-perturbation transcriptional responses across large pools of cancer cell lines, enabling deep characterization of shared and context-specific responses.
- James M. McFarland
- , Brenton R. Paolella
- & Aviad Tsherniak
-
Article
| Open Access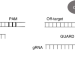 CRISPR GUARD protects off-target sites from Cas9 nuclease activity using short guide RNAs
CRISPR GUARD protects off-target sites from Cas9 nuclease activity using short guide RNAs
Off-target editing remains a concern for therapeutic applications of CRISPR-Cas9. Here the authors present CRISPR GUARD, which uses very short non-cleaving gRNAs to prevent editing at off-target sites.
- Matthew A. Coelho
- , Etienne De Braekeleer
- & Benjamin J. M. Taylor
-
Article
| Open Access Testing and controlling for horizontal pleiotropy with probabilistic Mendelian randomization in transcriptome-wide association studies
Testing and controlling for horizontal pleiotropy with probabilistic Mendelian randomization in transcriptome-wide association studies
Transcriptome-wide association studies integrate GWAS and transcriptome data to examine the molecular mechanisms underlying disease etiology. Here the authors present PMR-Egger, a powerful TWAS method based on probabilistic Mendelian Randomization.
- Zhongshang Yuan
- , Huanhuan Zhu
- & Xiang Zhou
-
Article
| Open Access A rare variant of African ancestry activates 8q24 lncRNA hub by modulating cancer associated enhancer
A rare variant of African ancestry activates 8q24 lncRNA hub by modulating cancer associated enhancer
Genetic variants on chromosome 8q24 are associated with prostate cancer risk in men of African ancestry. Here the authors show that one of these variants, rs72725854 alters the enhancer activity in its region, which upon androgen stimulation, activates multiple oncogenic lncRNAs and c-myc.
- Kaivalya Walavalkar
- , Bharath Saravanan
- & Dimple Notani
-
Article
| Open Access High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening
High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening
Reliable, multiplexed gene editing to uncover synergies between targets remains challenging. Here, the authors engineer AsCas12a and the crRNA to improve double knockout for synthetic sick/lethal interaction genetic screening.
- Rodrigo A. Gier
- , Krista A. Budinich
- & Junwei Shi
-
Article
| Open Access Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells
Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells
T cells are a major cell type involved in systemic lupus erythematosus (SLE). Here, the authors use promoter capture-C and ATAC-seq in human follicular T helper cells to identify SLE genes distant from GWAS loci (via 3D interaction) and validate the function of key regulatory elements and genes in vitro.
- Chun Su
- , Matthew E. Johnson
- & Andrew D. Wells
-
Article
| Open Access Germline de novo mutation rates on exons versus introns in humans
Germline de novo mutation rates on exons versus introns in humans
Evidence that somatic mutation rates in introns exceed those in exons challenges the molecular evolution tenet that mutation rate and sequence function are independent. Here, authors analyze germline de novo mutations and reveal no evidence for mutation rate differences between exons and introns.
- Miguel Rodriguez-Galindo
- , Sònia Casillas
- & Antonio Barbadilla
-
Article
| Open Access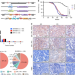 NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation
NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation
BRCA1 mutation carriers have higher chances of developing triple-negative breast cancer (TNBC). Here, the authors use the Sleeping Beauty mutagenesis system in Brca1 deficient mice and identify 169 putative driver genes, of which NOTCH1 accelerates TNBC formation through promoting epithelial-mesenchymal transition and cell cycle progression.
- Kai Miao
- , Josh Haipeng Lei
- & Chu-Xia Deng
-
Article
| Open Access RSPO3 impacts body fat distribution and regulates adipose cell biology in vitro
RSPO3 impacts body fat distribution and regulates adipose cell biology in vitro
Genetic variants at the RSPO3 locus are associated with waist-to-hip ratio (adjusted for BMI). Here, Loh et al. describe two independent RSPO3 signals that associate with body fat distribution, perform fine-mapping and explore the function of RSPO3 in human adipocyte biology and body fat distribution in zebrafish
- Nellie Y. Loh
- , James E. N. Minchin
- & Constantinos Christodoulides
-
Article
| Open Access A genome-wide gain-of-function screen identifies CDKN2C as a HBV host factor
A genome-wide gain-of-function screen identifies CDKN2C as a HBV host factor
Here the authors perform a gain-of-function screen and identify CDKN2C as a host factor for HBV replication, inducing cell cycle arrest and expression of HBV transcription enhancers. CDKN2C expression correlates with disease progression suggesting a potential role in HBV-induced liver disease.
- Carla Eller
- , Laura Heydmann
- & Thomas F. Baumert
-
Article
| Open Access Genomic adaptations to aquatic and aerial life in mayflies and the origin of insect wings
Genomic adaptations to aquatic and aerial life in mayflies and the origin of insect wings
Genomic studies of paleopteran insects, such as mayflies, are needed to reconstruct early insect evolution. Here, Almudi and colleagues present the genome of the mayfly Cloeon dipterum and use transcriptomics to characterize its adaptations to distinct habitats and the origin of insect wings.
- Isabel Almudi
- , Joel Vizueta
- & Fernando Casares
-
Article
| Open Access Evolutionary conserved NSL complex/BRD4 axis controls transcription activation via histone acetylation
Evolutionary conserved NSL complex/BRD4 axis controls transcription activation via histone acetylation
The MOF acetyltransferase-containing Non-Specific Lethal (NSL) complex is a broad transcription regulator and haploinsufficiency of its KANSL1 subunit results in the Koolen-de Vries syndrome in humans. Here, the authors identify the BET protein BRD4 as evolutionary conserved co-factor of the NSL complex and provide evidence that NSL-deposited histone acetylation induces BRD4 recruitment for transcription of constitutively active genes.
- Aline Gaub
- , Bilal N. Sheikh
- & Asifa Akhtar
-
Article
| Open Access FAM13A affects body fat distribution and adipocyte function
FAM13A affects body fat distribution and adipocyte function
Genetic variants in the FAM13A locus have been associated with anthropometric and glycemic traits. Here, using fine-mapping, in vitro knockdown studies in pre-adipocytes and in vivo knockout in mice, the authors show that FAM13A is involved in regulating fat distribution and metabolic traits.
- Mohsen Fathzadeh
- , Jiehan Li
- & Joshua W. Knowles
-
Article
| Open Access Abundance of conserved CRISPR-Cas9 target sites within the highly polymorphic genomes of Anopheles and Aedes mosquitoes
Abundance of conserved CRISPR-Cas9 target sites within the highly polymorphic genomes of Anopheles and Aedes mosquitoes
Genetic variation in natural populations could represent gene drive resistant alleles, preventing successful application for population management. Here the authors survey 1280 genomes from three mosquito species and concludes natural variation will not be detrimental to deploying gene drive technology.
- Hanno Schmidt
- , Travis C. Collier
- & Gregory C. Lanzaro
-
Article
| Open Access Nucleosome positioning stability is a modulator of germline mutation rate variation across the human genome
Nucleosome positioning stability is a modulator of germline mutation rate variation across the human genome
Nucleosome organization has been suggested to affect local mutation rates in the genome. Here, the authors analyse data on >300,000 human de novo mutations and high-resolution nucleosome maps and provide evidence that nucleosome positioning stability modulates germline mutation rate variation across the human genome.
- Cai Li
- & Nicholas M. Luscombe
-
Article
| Open Access Prioritizing disease and trait causal variants at the TNFAIP3 locus using functional and genomic features
Prioritizing disease and trait causal variants at the TNFAIP3 locus using functional and genomic features
While genome-wide association studies have yielded thousands of trait-associated loci, identifying causal variants remains challenging. Here, the authors perform seven genomics assays in various cell types to prioritize genetic variants in the TNFAIP3 locus, and report high-priority variants within disease-associated haplotypes.
- John P. Ray
- , Carl G. de Boer
- & Nir Hacohen
-
Article
| Open Access Gene clustering and copy number variation in alkaloid metabolic pathways of opium poppy
Gene clustering and copy number variation in alkaloid metabolic pathways of opium poppy
The opium poppy has been a source of painkilling drugs synthesized by the benzylisoquinoline alkaloid pathway. Here, the authors report an improved genome assembly and reveal gene clustering and copy number variation in alkaloid metabolic pathways.
- Qiushi Li
- , Sukanya Ramasamy
- & Sam Yeaman
-
Article
| Open Access Genome-wide association and multi-omic analyses reveal ACTN2 as a gene linked to heart failure
Genome-wide association and multi-omic analyses reveal ACTN2 as a gene linked to heart failure
Heart failure has a heterogeneous etiology and the genetic underpinnings are not well understood. Here, Arvanitis et al. perform GWAS meta-analysis including 10,976 heart failure cases and 437,573 controls, identify new loci near ABO and ACTN2 and show that deletion of a ACTN2 enhancer leads to reduced ACTN2 expression in differentiating cardiomyocytes.
- Marios Arvanitis
- , Emmanouil Tampakakis
- & Alexis Battle
-
Article
| Open Access In situ dissection of domain boundaries affect genome topology and gene transcription in Drosophila
In situ dissection of domain boundaries affect genome topology and gene transcription in Drosophila
In Drosophila, transcription is thought to be required for TAD formation, while the role of architectural proteins remains controversial. Here, the authors find that deletion of domain boundaries at the fly Notch locus results in TAD fusion and long-range topological defects, loss of architectural protein and RNA Pol II chromatin binding, and transcription defects.
- Rodrigo G. Arzate-Mejía
- , Angel Josué Cerecedo-Castillo
- & Félix Recillas-Targa
-
Article
| Open Access Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression
Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression
Studying the genetic effects on early stages of human development is challenging due to a scarcity of biological material. Here, the authors utilise induced pluripotent stem cells from 125 donors to track gene expression changes and expression quantitative trait loci at single cell resolution during in vitro endoderm differentiation.
- Anna S. E. Cuomo
- , Daniel D. Seaton
- & Oliver Stegle
-
Article
| Open Access De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences
De novo emergence of adaptive membrane proteins from thymine-rich genomic sequences
There is increasing evidence that protein-coding genes can emerge de novo from noncoding genomic regions. Vakirlis et al. propose that sequences encoding transmembrane polypeptides can emerge de novo in thymine-rich genomic regions and provide organisms with fitness benefits.
- Nikolaos Vakirlis
- , Omer Acar
- & Anne-Ruxandra Carvunis
-
Article
| Open Access Genetic screens in isogenic mammalian cell lines without single cell cloning
Genetic screens in isogenic mammalian cell lines without single cell cloning
Isogenic pairs of cell lines are powerful tools but time-consuming to generate. Here the authors conduct genome-wide genetic interactions screens of ‘anchor’ genes with SaCas9 and SpCas9.
- Peter C. DeWeirdt
- , Annabel K. Sangree
- & John G. Doench
-
Article
| Open Access A cancer rainbow mouse for visualizing the functional genomics of oncogenic clonal expansion
A cancer rainbow mouse for visualizing the functional genomics of oncogenic clonal expansion
Pre-malignant cells harbouring oncogenic mutations can populate and spread throughout a tissue. Here, using a rainbow mouse system, the authors explore how clonal expansion in the mouse intestine might explain high levels of intra-tumoural heterogeneity observed in the disease.
- Peter G. Boone
- , Lauren K. Rochelle
- & Joshua C. Snyder
-
Article
| Open Access Full-length transcriptome reconstruction reveals a large diversity of RNA and protein isoforms in rat hippocampus
Full-length transcriptome reconstruction reveals a large diversity of RNA and protein isoforms in rat hippocampus
It is challenging to characterize diverse transcript isoforms by short-read sequencing. Here the authors report full-length transcriptomes in rat hippocampus by hybrid-sequencing, predict isoform-specific translational status, and reconstruct open reading frames validated by mass spectrometry.
- Xi Wang
- , Xintian You
- & Wei Chen
-
Article
| Open Access GWAS for systemic sclerosis identifies multiple risk loci and highlights fibrotic and vasculopathy pathways
GWAS for systemic sclerosis identifies multiple risk loci and highlights fibrotic and vasculopathy pathways
Systemic sclerosis (SSc) is a heterogeneous chronic autoimmune disease that affects the connective tissue. Here, López-Isac et al. identify 13 new risk loci for SSc as well as loci specific for limited cutaneous and diffuse SSc and, defining credible sets and performing functional annotation, highlight key pathways and cell types for SSc.
- Elena López-Isac
- , Marialbert Acosta-Herrera
- & Javier Martin
-
Article
| Open Access Highly structured homolog pairing reflects functional organization of the Drosophila genome
Highly structured homolog pairing reflects functional organization of the Drosophila genome
Trans-homolog interactions, such as homolog pairing, are highly structured and associated with gene function in Drosophila cells. Here, the authors use haplotype-resolved Hi-C to identify genome-wide trans-homolog interactions in a Drosophila hybrid cell line and investigate their patterns and functional roles.
- Jumana AlHaj Abed
- , Jelena Erceg
- & C.-ting Wu
-
Article
| Open Access BORDER proteins protect expression of neighboring genes by promoting 3′ Pol II pausing in plants
BORDER proteins protect expression of neighboring genes by promoting 3′ Pol II pausing in plants
In plants, 3′ Pol II pausing helps prevent transcriptional interference. Here the authors provide evidence that BORDER proteins are enriched in intergenic regions and inhibit transcriptional interference between closely spaced genes by preventing RNA polymerase from intruding into the promoters of nearby downstream genes.
- Xuhong Yu
- , Pascal G. P. Martin
- & Scott D. Michaels
-
Article
| Open Access Identification of significant chromatin contacts from HiChIP data by FitHiChIP
Identification of significant chromatin contacts from HiChIP data by FitHiChIP
HiChIP/PLAC-seq assay is popular for profiling 3D genome interactions among regulatory elements at kilobase resolution. Here the authors describe FitHiChIP an empirical null-based, flexible computational method for statistical significance estimation and loop calling from HiChIP data.
- Sourya Bhattacharyya
- , Vivek Chandra
- & Ferhat Ay
-
Article
| Open Access Evolutionary and functional impact of common polymorphic inversions in the human genome
Evolutionary and functional impact of common polymorphic inversions in the human genome
Inversions are a little-studied type of genomic variation that could contribute to phenotypic traits. Here the authors characterize 45 common polymorphic inversions in human populations and investigate their evolutionary and functional impact.
- Carla Giner-Delgado
- , Sergi Villatoro
- & Mario Cáceres
-
Article
| Open Access In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene
In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene
Although many neuropsychiatric risk genes are known to contribute to epigenetic regulation of gene expression, very little is known about specific chromatin-associated mechanisms that govern the formation and maintenance of neuronal connectivity. Here, the authors report that transcallosal connectivity is critically dependent on C11orf46/ARL14EP, a nuclear protein encoded in the chromosome 11p13 WAGR risk locus, and that RNA-guided epigenetic editing of hyperexpressed Sema6a gene promoters in C11orf46-knockdown neurons resulted in normalization of expression and rescue of transcallosal dysconnectivity via repressive chromatin remodeling.
- Cyril J. Peter
- , Atsushi Saito
- & Atsushi Kamiya
-
Article
| Open Access Mitigation of off-target toxicity in CRISPR-Cas9 screens for essential non-coding elements
Mitigation of off-target toxicity in CRISPR-Cas9 screens for essential non-coding elements
Off-target effects in CRISPR screens for essential regulatory elements have not been systematically evaluated. Here the authors find Cas9 nuclease, CRISPRi/a each have distinct off-target effects, and that these can be accurately identified and removed using the GuideScan sgRNA specificity score.
- Josh Tycko
- , Michael Wainberg
- & Michael C. Bassik
-
Article
| Open Access Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution
Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution
Interpreting genetic variation in the noncoding genome remains challenging, with functional effects difficult to predict. Here, the authors perform saturation mutagenesis combined with massively parallel reporter assays for 20 disease-associated regulatory elements, quantifying the effects of over 30,000 variants.
- Martin Kircher
- , Chenling Xiong
- & Nadav Ahituv
-
Article
| Open Access Functional genetic variants can mediate their regulatory effects through alteration of transcription factor binding
Functional genetic variants can mediate their regulatory effects through alteration of transcription factor binding
Functional variants have been proposed to alter transcription factor binding. Here, the authors provide direct evidence that functional variants within the TBC1D4 gene, encoding an NFκB binding site, can alter transcription factor binding, and use CRISPR-Cas9 to reveal localization of the transcription factor to be the regulator of chromatin accessibility and p65 binding and ultimately TBC1D4 expression.
- Andrew D. Johnston
- , Claudia A. Simões-Pires
- & John M. Greally
-
Article
| Open Access MAPCap allows high-resolution detection and differential expression analysis of transcription start sites
MAPCap allows high-resolution detection and differential expression analysis of transcription start sites
The position, shape and number of transcription start sites (TSS) regulate gene expression. Here authors present MAPCap, a method for high-resolution detection and differential expression analysis of TSS, and apply MAPCap to early fly development, detecting stage and sex-specific promoter and enhancer activity.
- Vivek Bhardwaj
- , Giuseppe Semplicio
- & Asifa Akhtar

 LETR1 is a lymphatic endothelial-specific lncRNA governing cell proliferation and migration through KLF4 and SEMA3C
LETR1 is a lymphatic endothelial-specific lncRNA governing cell proliferation and migration through KLF4 and SEMA3C