Featured
-
-
Article
| Open Access Reprogramming mechanism dissection and trophoblast replacement application in monkey somatic cell nuclear transfer
Reprogramming mechanism dissection and trophoblast replacement application in monkey somatic cell nuclear transfer
Somatic cloning of rhesus monkey has not been successful until now. Here, authors report epigenetic abnormalities in SCNT embryos and placentas and develop a trophoblast replacement method that enables them to successful clone of a healthy male rhesus monkey.
- Zhaodi Liao
- , Jixiang Zhang
- & Qiang Sun
-
Article
| Open Access Epigenomic analysis of formalin-fixed paraffin-embedded samples by CUT&Tag
Epigenomic analysis of formalin-fixed paraffin-embedded samples by CUT&Tag
Conducting epigenomic studies on FFPE samples is traditionally challenging due to chromatin damage caused due to exposure to formaldehyde. Here, the authors show that an optimisation of their previous CUTAC method allows the production of high-resolution maps of regulatory elements from FFPE samples.
- Steven Henikoff
- , Jorja G. Henikoff
- & Eric C. Holland
-
Article
| Open Access SHIELD: a platform for high-throughput screening of barrier-type DNA elements in human cells
SHIELD: a platform for high-throughput screening of barrier-type DNA elements in human cells
Chromatin boundary elements are hard to define and characterize. Here the authors report Site-specific Heterochromatin Insertion of Elements at Lamina-associated Domains (SHIELD) for high-throughput screening of barrier-type DNA elements in human cells.
- Meng Zhang
- , Mary Elisabeth Ehmann
- & Huimin Zhao
-
Article
| Open Access Epigenome-wide association analysis of infant bronchiolitis severity: a multicenter prospective cohort study
Epigenome-wide association analysis of infant bronchiolitis severity: a multicenter prospective cohort study
DNA methylation patterns that are associated with disease can reveal genes involved in disease etiology. Here, the authors identify blood DNA methylation signatures that are associated with bronchiolitis severity and play important roles in tissues, cells, and pathways.
- Zhaozhong Zhu
- , Yijun Li
- & Kohei Hasegawa
-
Article
| Open Access A qPCR technology for direct quantification of methylation in untreated DNA
A qPCR technology for direct quantification of methylation in untreated DNA
Analysis of DNA methylation usually requires a chemical or an enzymatic pretreatment step. Here, the authors report a PCR-based technology for the detection of DNA methylation in untreated DNA, and present analytical and clinical results from methylation analysis of the MGMT promoter.
- Kamilla Kolding Bendixen
- , Maria Mindegaard
- & Rasmus Koefoed Petersen
-
Article
| Open Access Droplet-based bisulfite sequencing for high-throughput profiling of single-cell DNA methylomes
Droplet-based bisulfite sequencing for high-throughput profiling of single-cell DNA methylomes
Single-cell DNA methylomic studies offer high resolution to differentiate cell subsets based on their epigenomic features. Here, the authors demonstrate Drop-BS, a droplet-based single-cell bisulfite sequencing library preparation method, for DNA methylome profiling.
- Qiang Zhang
- , Sai Ma
- & Chang Lu
-
Article
| Open Access Roving methyltransferases generate a mosaic epigenetic landscape and influence evolution in Bacteroides fragilis group
Roving methyltransferases generate a mosaic epigenetic landscape and influence evolution in Bacteroides fragilis group
Here, Tisza, Dekker, and colleagues perform large scale analysis of genome methylation in the gut commensal and pathogen, Bacteroides fragilis group, revealing immense methyl motif diversity and evidence of widespread methyltransferase exchange among phages.
- Michael J. Tisza
- , Derek D. N. Smith
- & John P. Dekker
-
Article
| Open Access HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing
HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing
Limited work has been done on concurrent C-to-G and A-to-G base editing. Here the authors test how a number of chromatin-associated factors affect base editing and show that HMGN1 enhanced the efficiency; by fusing HMGN1 to GBE and ABE they develop a CRISPR-based dual-function A-to-G and C-to-G base editor (GGBE).
- Chao Yang
- , Zhenzhen Ma
- & Xueli Zhang
-
Article
| Open Access Low input capture Hi-C (liCHi-C) identifies promoter-enhancer interactions at high-resolution
Low input capture Hi-C (liCHi-C) identifies promoter-enhancer interactions at high-resolution
Here the authors present the low input capture Hi-C (liCHi-C) method, a cost-effective, flexible method to map and robustly compare promoter interactomes at high resolution. liCHi-C identifies new disease-associated genes and structural variants to ultimately illuminate their pathogenic effects.
- Laureano Tomás-Daza
- , Llorenç Rovirosa
- & Biola M. Javierre
-
Article
| Open Access Simultaneous profiling of histone modifications and DNA methylation via nanopore sequencing
Simultaneous profiling of histone modifications and DNA methylation via nanopore sequencing
The interplay between histone modifications and DNA methylation plays a crucial role in establishing and maintaining the epigenomic landscape. Here, the authors develop a nanopore sequencing based method for mapping histone modifications and DNA methylation from native, long, single DNA molecules.
- Xue Yue
- , Zhiyuan Xie
- & Yimeng Yin
-
Article
| Open Access High-throughput robust single-cell DNA methylation profiling with sciMETv2
High-throughput robust single-cell DNA methylation profiling with sciMETv2
Despite the importance of DNA methylation, accessible and high-throughput methods to profile methylation at the single-cell level are lacking. Here, the authors present sciMETv2, a high-throughput workflow that provides high-quality single-cell methylomes in a robust and simple workflow.
- Ruth V. Nichols
- , Brendan L. O’Connell
- & Andrew C. Adey
-
Article
| Open Access High-affinity chromodomains engineered for improved detection of histone methylation and enhanced CRISPR-based gene repression
High-affinity chromodomains engineered for improved detection of histone methylation and enhanced CRISPR-based gene repression
Engineered chromodomains are promising probes to analyze histone methylation. Here the authors designed high-affinity chromodomains for enhanced genome-wide binding analysis, live-cell imaging and ultra-potent CRISPR-based gene repression.
- G. Veggiani
- , R. Villaseñor
- & S. S. Sidhu
-
Article
| Open Access Learning the histone codes with large genomic windows and three-dimensional chromatin interactions using transformer
Learning the histone codes with large genomic windows and three-dimensional chromatin interactions using transformer
Existing deep learning-based approaches for the prediction of gene expression by histone modifications (HMs) can only focus on narrow and linear genomic regions around promoters. Here, the authors address these problems by developing a transformer-based deep learning architecture named Chromoformer.
- Dohoon Lee
- , Jeewon Yang
- & Sun Kim
-
Article
| Open Access Hi-TrAC reveals division of labor of transcription factors in organizing chromatin loops
Hi-TrAC reveals division of labor of transcription factors in organizing chromatin loops
It is currently not clear how architectural proteins orchestrate chromatin looping at different scales of genome organisation. Here the authors report Hi-TrAC as a proximity ligation-free method to profile genome-wide chromatin interactions at single nucleosome resolution among regulatory elements.
- Shuai Liu
- , Yaqiang Cao
- & Keji Zhao
-
Article
| Open Access Multidimensional chromatin profiling of zebrafish pancreas to uncover and investigate disease-relevant enhancers
Multidimensional chromatin profiling of zebrafish pancreas to uncover and investigate disease-relevant enhancers
Alterations in cis-regulatory elements (CREs) can contribute to pancreatic diseases. Here the authors combine chromatin profiling and interaction points with in vivo reporter assays in zebrafish to uncover functionally equivalent human CREs, helping to predict disease-relevant enhancers.
- Renata Bordeira-Carriço
- , Joana Teixeira
- & José Bessa
-
Article
| Open Access Cell-intrinsic Aryl Hydrocarbon Receptor signalling is required for the resolution of injury-induced colonic stem cells
Cell-intrinsic Aryl Hydrocarbon Receptor signalling is required for the resolution of injury-induced colonic stem cells
Rapid intestinal regeneration after injury is critical to maintain barrier integrity and homeostasis, but must be tightly controlled to prevent tumorigenesis. Here they show that the aryl hydrocarbon receptor is required to terminate the regenerative response after wound healing.
- Kathleen Shah
- , Muralidhara Rao Maradana
- & Brigitta Stockinger
-
Article
| Open Access A fast Myosin super enhancer dictates muscle fiber phenotype through competitive interactions with Myosin genes
A fast Myosin super enhancer dictates muscle fiber phenotype through competitive interactions with Myosin genes
The contractile properties of adult myofibers are shaped by their Myosin heavy chain isoform content. Here the authors show that a super enhancer controls the spatiotemporal expression of the genes at the fast myosin heavy chain locus by DNA looping and that this expression profile is recapitulated in a rainbow transgenic mouse model of the locus.
- Matthieu Dos Santos
- , Stéphanie Backer
- & Pascal Maire
-
Article
| Open Access Integration of single-cell transcriptomes and chromatin landscapes reveals regulatory programs driving pharyngeal organ development
Integration of single-cell transcriptomes and chromatin landscapes reveals regulatory programs driving pharyngeal organ development
The molecular basis and gene regulatory networks driving pharyngeal endoderm development remain poorly understood. Here the authors report single cell transcriptomic and chromatin landscapes to delineate regulatory programs driving this process and to define the immunodeficiency-associated developmental defects resulting from Foxn1 dysfunction.
- Margaret E. Magaletta
- , Macrina Lobo
- & René Maehr
-
Article
| Open Access Dynamic transcriptome and chromatin architecture in granulosa cells during chicken folliculogenesis
Dynamic transcriptome and chromatin architecture in granulosa cells during chicken folliculogenesis
The domestic chicken Gallus gallus domesticus is a classic model for the study of folliculogenesis. Here the authors integrate multi-omics analyses characterizing the dynamic transcriptome and chromatin architecture in granulosa cells during chicken folliculogenesis.
- Diyan Li
- , Chunyou Ning
- & Mingzhou Li
-
Article
| Open Access A compendium of chromatin contact maps reflecting regulation by chromatin remodelers in budding yeast
A compendium of chromatin contact maps reflecting regulation by chromatin remodelers in budding yeast
The effect of ATP-dependent chromatin remodelers on 3D genome organization has not been well studied. Here the authors employ in situ Hi-C with an auxin-inducible degron system to degrade chromatin remodelers in yeast to find that the 3D structure of chromatin collapses in their absence. The chromatin remodeling can modulate 3D architecture depending on chromosomal context and cell cycle stage.
- Hyelim Jo
- , Taemook Kim
- & Daeyoup Lee
-
Article
| Open Access Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9
Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9
The causal relationship between DNA demethylation and gene expression regulation has not yet been fully resolved. Here the authors develop a nuclease-dead Cas9 (dCas9) and gRNA site-specific targeting approach to physically block DNA methylation at specific promoters to cause DNA demethylation in cells and tackle this question.
- Daniel M. Sapozhnikov
- & Moshe Szyf
-
Article
| Open Access EpiScanpy: integrated single-cell epigenomic analysis
EpiScanpy: integrated single-cell epigenomic analysis
The authors present epiScanpy: a computational framework for the analysis of single-cell epigenomic data, both ATAC-seq and DNA methylation data, with examples for clustering, cell type identification, trajectory learning and atlas integration - and show its performance in distinguishing cell types.
- Anna Danese
- , Maria L. Richter
- & Maria Colomé-Tatché
-
Article
| Open Access Mesomelic dysplasias associated with the HOXD locus are caused by regulatory reallocations
Mesomelic dysplasias associated with the HOXD locus are caused by regulatory reallocations
Mesomelic dysplasia, a severe shortening and bending of the limb, has been linked to rearrangements in the HoxD cluster in humans and mice. Here the authors engineer a 1 Mb inversion including the HoxD gene cluster and use this model to provide a mechanistic framework to understand and unify the molecular origins of human mesomelic dysplasia associated with 2q31.
- Christopher Chase Bolt
- , Lucille Lopez-Delisle
- & Denis Duboule
-
Article
| Open Access HIRA stabilizes skeletal muscle lineage identity
HIRA stabilizes skeletal muscle lineage identity
The epigenetic mechanisms coordinating the maintenance of adult cellular lineages remain poorly understood. Here the authors demonstrate that HIRA, a H3.3 histone chaperone, establishes the chromatin landscape required for skeletal muscle cell identity.
- Joana Esteves de Lima
- , Reem Bou Akar
- & Frédéric Relaix
-
Article
| Open Access Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells
Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells
Histone variant H2A.Z has been suggested to contribute to the regulation of promoter accessibility. Here, the authors present high-depth maps of the position and accessibility of H2A.Z-containing nucleosomes for human Pol II promoters and provide evidence that H2A.Z has multiple and distinct roles in regulating gene expression dependent upon its location in a promoter.
- Lauren Cole
- , Sebastian Kurscheid
- & David J. Tremethick
-
Article
| Open Access DNA methylation predicts age and provides insight into exceptional longevity of bats
DNA methylation predicts age and provides insight into exceptional longevity of bats
DNA methylation profiles from 26 bat species accurately predicts chronological age, while longevity-related methylation patterns across the genome suggest that bat longevity results from augmented immune response and cancer suppression.
- Gerald S. Wilkinson
- , Danielle M. Adams
- & Steve Horvath
-
Article
| Open Access Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos
Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos
Extensive epigenetic reprogramming occurs during preimplantation embryo development. Here the authors develop a single cell multiomics sequencing technology that enables profiling of genome-wide chromatin accessibility, DNA methylation and RNA expression in the same individual cell and apply this method to study mouse preimplantation embryos.
- Yang Wang
- , Peng Yuan
- & Liying Yan
-
Article
| Open Access Non-CG methylation and multiple histone profiles associate child abuse with immune and small GTPase dysregulation
Non-CG methylation and multiple histone profiles associate child abuse with immune and small GTPase dysregulation
Early-life adversity is thought to increase the risk of psychopathology through epigenetic mechanisms. Here, the authors profile 6 histone marks, chromatin states and DNA methylation in the lateral amygdala in subjects with a history of early-life adversity.
- Pierre-Eric Lutz
- , Marc-Aurèle Chay
- & Gustavo Turecki
-
Article
| Open Access The genome-wide impact of trisomy 21 on DNA methylation and its implications for hematopoiesis
The genome-wide impact of trisomy 21 on DNA methylation and its implications for hematopoiesis
Down syndrome has a high co-morbidity with immune and hematopoietic disorders. Here, the authors perform an epigenome-wide association study in newborns with and without Down syndrome to find differential methylation across the genome, including in hematopoietic regulators RUNX1 and FLI1.
- Ivo S. Muskens
- , Shaobo Li
- & Adam J. de Smith
-
Article
| Open Access An all-to-all approach to the identification of sequence-specific readers for epigenetic DNA modifications on cytosine
An all-to-all approach to the identification of sequence-specific readers for epigenetic DNA modifications on cytosine
Identifying readers of epigenetic marks is a critical step for understanding the role of epigenetic marks in biology. Here, the authors applied DAPPL, an all-to-all approach to profile the interactions between TFs and epigenetic modified DNA libraries.
- Guang Song
- , Guohua Wang
- & Heng Zhu
-
Article
| Open Access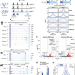 Conserved regulatory logic at accessible and inaccessible chromatin during the acute inflammatory response in mammals
Conserved regulatory logic at accessible and inaccessible chromatin during the acute inflammatory response in mammals
Genetic elements that control inflammatory gene expression are not fully elucidated. Here the authors conduct a multi-species analysis of chromatin landscape and NF-κB binding in response to the proinflammatory cytokine TNFα, finding that conserved NF-κB bound regions are linked to enhancer activity and disease.
- Azad Alizada
- , Nadiya Khyzha
- & Michael D. Wilson
-
Article
| Open Access GATA2 regulates mast cell identity and responsiveness to antigenic stimulation by promoting chromatin remodeling at super-enhancers
GATA2 regulates mast cell identity and responsiveness to antigenic stimulation by promoting chromatin remodeling at super-enhancers
Mast cells are critical effectors of allergic inflammation and protection against parasitic infections. Here the authors demonstrate that GATA2 promotes chromatin accessibility at the super-enhancers of mast cell identity genes and primes both typical and super-enhancers at genes that respond to antigenic stimulation.
- Yapeng Li
- , Junfeng Gao
- & Hua Huang
-
Article
| Open Access BET inhibition disrupts transcription but retains enhancer-promoter contact
BET inhibition disrupts transcription but retains enhancer-promoter contact
The role of BRD4 and Mediator in regulating enhancer-promoter interactions is poorly understood. Here the authors find that treatment with BET inhibitors or pharmacological degradation of BRD4 disrupts transcription while having very little effect on enhancer-promoter interactions.
- Nicholas T. Crump
- , Erica Ballabio
- & Thomas A. Milne
-
Article
| Open Access Transcription shapes genome-wide histone acetylation patterns
Transcription shapes genome-wide histone acetylation patterns
Histone acetylation is a ubiquitous hallmark of transcription. Here the authors provide evidence that the majority of histone acetylation is dependent on transcription, specifically due to the requirement of RNAPII for the recruitment and activity of histone acetyltransferases.
- Benjamin J. E. Martin
- , Julie Brind’Amour
- & LeAnn J. Howe
-
Article
| Open Access Order and stochasticity in the folding of individual Drosophila genomes
Order and stochasticity in the folding of individual Drosophila genomes
Genomes are partitioned into topologically associating domains (TADs). Here the authors present single-nucleus Hi-C maps in Drosophila at 10 kb resolution, demonstrating the presence of chromatin compartments in individual nuclei, and partitioning of the genome into non-hierarchical TADs at the scale of 100 kb, which resembles population TAD profiles.
- Sergey V. Ulianov
- , Vlada V. Zakharova
- & Sergey V. Razin
-
Article
| Open Access Hydroxamic acid-modified peptide microarrays for profiling isozyme-selective interactions and inhibition of histone deacetylases
Hydroxamic acid-modified peptide microarrays for profiling isozyme-selective interactions and inhibition of histone deacetylases
Current histone microarrays cannot be used to directly study the transient interactions of histone deacetylases (HDACs). Here, the authors show that hydroxamic acid-modified microarrays can capture HDACs, provide insights into their substrate specificity, and serve to develop peptide inhibitors.
- Carlos Moreno-Yruela
- , Michael Bæk
- & Christian A. Olsen
-
Article
| Open Access Primary effusion lymphoma enhancer connectome links super-enhancers to dependency factors
Primary effusion lymphoma enhancer connectome links super-enhancers to dependency factors
Primary effusion lymphoma (PEL) has a very poor prognosis. Here, the authors perform H3K27ac HiChIP in PEL cells and generate the PEL enhancer connectome, linking enhancers and promoters in PEL, as well as super-enhancers to dependency factors.
- Chong Wang
- , Luyao Zhang
- & Bo Zhao
-
Article
| Open Access An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas
An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas
Strong transgene suppression has been observed in Chlamydomonas reinhardtii, but the underlying mechanism is unknown. Here, the authors identify a sirtuin-type histone deacetylase that selectively acts on transgenic DNA to repress gene expression by assembling a repressive chromatin structure composed of deacetylated histones.
- Juliane Neupert
- , Sean D. Gallaher
- & Ralph Bock
-
Article
| Open Access SAMMY-seq reveals early alteration of heterochromatin and deregulation of bivalent genes in Hutchinson-Gilford Progeria Syndrome
SAMMY-seq reveals early alteration of heterochromatin and deregulation of bivalent genes in Hutchinson-Gilford Progeria Syndrome
Hutchinson-Gilford progeria syndrome is a genetic disease where an aberrant form of Lamin A disrupts chromatin by interfering with lamina associated domains. Here, the authors present the SAMMY-seq, a method for genome-wide characterization of heterochromatin dynamics and detect early stage alterations of heterochromatin structure in progeria primary fibroblasts, accompained by Polycomb dysfunctions.
- Endre Sebestyén
- , Fabrizia Marullo
- & Chiara Lanzuolo
-
Article
| Open Access Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence
Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence
Senescence is a state of stable proliferative arrest. Here, the authors perform Hi-C analysis on oncogenic RAS-induced senescence in human fibroblasts and characterize the changes in the 3D genome folding associated with the senescence-specific gene expression profile, which are mediated in part through cohesin redistribution on chromatin.
- Ioana Olan
- , Aled J. Parry
- & Masashi Narita
-
Article
| Open Access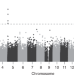 Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies methylation changes in AHRR
Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies methylation changes in AHRR
PTSD has been associated with DNA methylation of specific loci in the genome, but studies have been limited by small sample sizes. Here, the authors perform a meta-analysis of DNA methylation data from 10 different cohorts and identify CpGs in AHRR that are associated with PTSD.
- Alicia K. Smith
- , Andrew Ratanatharathorn
- & Caroline M. Nievergelt
-
Article
| Open Access Detection of haplotype-dependent allele-specific DNA methylation in WGBS data
Detection of haplotype-dependent allele-specific DNA methylation in WGBS data
Allele-specific measurements can reveal differences in DNA methylation between homologous alleles associated with changes in genetic sequence. Here, the authors develop a method for detecting allele specific methylation events within haplotypes of linked SNPs, compare it with existing methods, and show it identifies haplotypes for which the genetic variant carries significant information about the methylation state of the allele of origin.
- J. Abante
- , Y. Fang
- & J. Goutsias
-
Article
| Open Access An embryonic stem cell-specific heterochromatin state promotes core histone exchange in the absence of DNA accessibility
An embryonic stem cell-specific heterochromatin state promotes core histone exchange in the absence of DNA accessibility
Nucleosome turnover concomitant with incorporation of the histone variant H3.3 is a hallmark of regulatory regions in the animal genome. Here, the authors demonstrate that fast histone turnover and H3.3 incorporation defines a dynamic heterochromatin state in pluripotent stem cells.
- Carmen Navarro
- , Jing Lyu
- & Simon J. Elsässer
-
Article
| Open Access Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution
Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution
Claudin-low tumors are a rare aggressive subtype of breast cancers. In this study, the authors use a multiomics approach to demonstrate that these tumors are heterogeneous and comprise three main subgroups that emerge from different evolutionary processes.
- Roxane M. Pommier
- , Amélien Sanlaville
- & Alain Puisieux
-
Article
| Open Access Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells
Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells
T cells are a major cell type involved in systemic lupus erythematosus (SLE). Here, the authors use promoter capture-C and ATAC-seq in human follicular T helper cells to identify SLE genes distant from GWAS loci (via 3D interaction) and validate the function of key regulatory elements and genes in vitro.
- Chun Su
- , Matthew E. Johnson
- & Andrew D. Wells
-
Article
| Open Access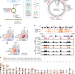 Epigenetic specifications of host chromosome docking sites for latent Epstein-Barr virus
Epigenetic specifications of host chromosome docking sites for latent Epstein-Barr virus
Epstein-Barr virus (EBV) episomes tether to the host chromosome via EBNA1. Here, using circular chromosome conformation capture (4C), Kim et al. identify attachment sites and show that EBV episomes preferentially associate with transcriptionally silenced genes in Burkitt lymphoma cells.
- Kyoung-Dong Kim
- , Hideki Tanizawa
- & Paul M. Lieberman
-
Article
| Open Access Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing
Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing
Tissues-specific gene expression requires coordinated cis-regulatory elements. Here the authors use dCas9-based enhancer targeting to remodel local epigenetic landscapes and activate or inactive transcription.
- Kailong Li
- , Yuxuan Liu
- & Jian Xu
-
Article
| Open Access Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes
Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes
N6-methyladenosine (m6A) and N6,2′-O-dimethyladenosine (m6Am) are eukaryotic mRNA modifications. Here the authors develop m6A-Crosslinking-Exonuclease-sequencing to map quantitative methylome changes at single-base-resolution after individually knocking out each known methyltransferase or demethylase.
- Casslynn W. Q. Koh
- , Yeek Teck Goh
- & W. S. Sho Goh
-
Article
| Open Access The epigenomic landscape of transposable elements across normal human development and anatomy
The epigenomic landscape of transposable elements across normal human development and anatomy
Although most are silenced, certain transposable elements (TEs) have been co-opted by the host. Here, the authors quantify the epigenomic status of TEs using data from the Roadmap Epigenomics Project, provide a systematic profile of TE activity across normal human tissues and development, finding that TEs encompass a quarter of the human regulatory epigenome, with 47% of TEs in regulatory states.
- Erica C. Pehrsson
- , Mayank N. K. Choudhary
- & Ting Wang

 Polycomb-mediated silencing of miR-8 is required for maintenance of intestinal stemness in Drosophila melanogaster
Polycomb-mediated silencing of miR-8 is required for maintenance of intestinal stemness in Drosophila melanogaster