Abstract
There is increasing interest in the development of noninvasive diagnostic methods for early cancer detection, to improve the survival rate and quality of life of cancer patients. Identification of volatile metabolic compounds may provide an approach for noninvasive early diagnosis of malignant diseases. Here we analyzed the volatile metabolic signature of human breast cancer cell lines versus normal human mammary cells. Volatile compounds in the headspace of conditioned culture medium were directly fingerprinted by secondary electrospray ionization-mass spectrometry. The mass spectra were subsequently treated statistically to identify discriminating features between normal vs. cancerous cell types. We were able to classify different samples by using feature selection followed by principal component analysis (PCA). Additionally, high-resolution mass spectrometry allowed us to propose their chemical structures for some of the most discriminating molecules. We conclude that cancerous cells can release a characteristic odor whose constituents may be used as disease markers.
Similar content being viewed by others
Introduction
Breast cancer is the most prevalent malignant disease threatening women worldwide. The survival rate can be greatly improved due to better diagnostic screening methods for early-stage disease with reliable biomarkers1. However, the current situation is that reliable metabolic biomarkers of breast cancer are still lacking. Therefore, efforts are increasingly made to detect breast cancer based on a panel of small molecules derived from the metabolic profiles of samples such as urine, blood or tissue2,3. Gas chromatography-mass spectrometry (GC-MS)4 and liquid chromatography-mass spectrometry (LC-MS) have been routinely used for metabolic fingerprinting analysis5. However, analyzing urine or blood with these routine methods involves time-consuming sample preparation and separation procedures, which are difficult to apply for clinical diagnosis. Analyzing a readily accessible and informative sample with a real-time method would be a promising approach for early detection of breast cancer. Recently, a number of studies have suggested a correlation between volatile compounds released by the human body and the health status of an individual6. Interestingly, a number of studies suggest that dogs are able to identify cancerous versus healthy tissues simply by sniffing them7,8,9. This indicates that some specific volatile compounds may originate from cancer cells as a result of their abnormal metabolism.
Similarly to previous studies10,11, to test this hypothesis, we chose the supernatant of culture media as samples. This offers the advantage that there are no subsequent interferences along the excretion pathway, making it possible to provide an unaltered metabolite pattern of cellular origin12. We analyzed the volatiles released from breast cancer cells versus healthy mammary cells by real-time secondary electrospray ionization mass spectrometry13,14,15,16,17,18 Subsequent statistical analysis revealed a panel of molecules that allowed the discrimination of the different groups. Furthermore, high resolution mass spectrometry was applied to give some tentative peak assignments to some of the most significant molecules. These breast cancer specific metabolic markers demonstrate the potential use of volatile metabolites in early diagnosis of malignant diseases.
Results
Real-time analysis of volatiles and feature selection between MDA-MB-231 vs. HMLE
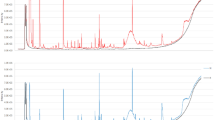
Figure 1a shows the real-time intensity vs. scan numbers (i.e. time) of some compounds detected in the headspace of the culture media after cell growing. It illustrates a typical series of headspace measurements recorded for two samples of culture medium of the normal cell line (HMLE) and the breast cancer cell line (MDA-MB-231) in both positive and negative ion modes. For each compound and for a given cell culture medium, the signal increased above the background level during each headspace sampling. Each sample provided six consecutive headspace technical replicates with satisfactory repeatability. This clearly suggests that the discriminating signals stem from metabolites in the culture medium.
(a) Typical series of real-time headspace analyses recorded from culture medium of normal and cancerous cells. The relative intensity vs. scan numbers of four compounds is shown (m/z 114.0914, m/z 185.1187 in positive mode and m/z 155.108, m/z 211.1341 in negative mode, respectively). From each culture, six consecutive headspace samples were analyzed and they had a comparable height within each sample, but the intensities were different between various samples. (b) Box plots of two exemplarily features (in both negative mode and positive mode) that show significantly different molecular profiles. The asterisks on the top of the figures represent p-values (one asterisk: P < 0.05, two asterisks: P < 0.01, three asterisks: P < 0.005), which were computed using a Mann–Whitney U test. The peaks on the right show zoomed views of the corresponding mass spectra.
The Mann–Whitney U test and further feature selection (see methods section) led to 16 compounds (9 in negative ion mode and 7 in positive ion mode). These were emitted with statistical significance by the cancer cells as compared to the normal ones. Because we kept the pH value of the culture medium for the normal and breast cancer cells line approximately the same, we conclude that the significant differences found for both cells types could not be attributed to this effect11.
Interestingly enough, the 7 compound detected in positive ion mode and the 9 volatiles detected in negative ion mode, were systematically found in the cancer cells decreased and increased, respectively. This is illustrated in Figure 1b for the four representative ions of Figure 1a. The box plots provide an overview of the differences found for all the normal and cancer cells measured. The right panels display the zoomed views of the corresponding mass spectra in the mass window of interest. In summary, the univariate analysis results suggest that the culture medium headspace of normal cells and of breast cancer cells have their unique volatile fingerprints. This notion was strengthened through further multivariate analysis.
PCA analysis and class prediction for MDA-MB-231 vs. HMLE
As described above, the 16 m/z variables resulting from the Mann–Whitney U test were retained for PCA and the results are illustrated in Figure 2, which shows the score plot for the first two principal components. Clearly, normal cells and cancer cells occupy their own space (i.e. positive (negative) values for PC1 in cancer (normal) cells), reinforcing the notion of the difference in the culture medium headspace for normal vs. cancer cells.
PCA scatter plot of the headspace of medium culture for normal mammary cells (10 biological replicates) and breast cancer cells (10 biological replicates) and loadings for PC1 in the PCA model.
Negative loadings indicate an increase in normal cells while positive loadings indicate an increase in cancer cells.
Finally, we attempted to predict cell class based on the pattern imparted by the selected 16 volatile molecules by a k-nearest-neighbor (KNN; k = 1, Euclidean distance) algorithm in a leave-one-out cross validation. The overall correct classification score was 90% (i.e. one cancer and one normal sample were misclassified) indicating that the selected volatile fingerprint may be useful to predict MDA-MB-231 vs. HMLE.
Verifications in different human breast cancer cell lines
To further verify the 16 selected volatile molecules, we confirmed the presence of these molecules in the headspace of the culture medium from two types of less aggressive (T47D) and medium aggressive (SKBR-3) breast cancer cell lines(see supporting Figure S1, S2). From the box plot, we can observe that in negative ion mode, in all three types of breast cancer cell lines, the 9 selected molecules with significant differences consistently showed higher intensity than that in the normal cell line, with the highest concentration in the T47D and the lowest in the SKBR-3 cell lines (see supporting Figure S1). Whereas, in positive ion mode, we can observe that the 7 selected molecules with significant differences showed an obvious decreasing trend in the cancer cell lines (see supporting Figure S2). Furthermore, we measured the proliferation rate of the three types of breast cancer cell lines to figure out the reason for concentration variations in the 16 significantly different peaks. The result (see supporting Figure S3) showed that the SKBR-3 cells grew the fastest while the T47D cells grew the slowest, just opposite to the concentration change in significantly different molecules detected in negative ion mode.
Tentative molecule identification
Elemental compositions for the 16 molecules which showed significant difference between normal and cancer cells were determined, using high resolution, accurate mass measurements and isotopic patterns (Table 1). In negative ion mode, the 9 significantly different compounds were all in the category of fatty acids. In positive ion mode, most of the 7 discriminate peaks were nutrients from culture medium.
Discussion
In positive ion mode, some of the 7 compounds showing significant difference may be in the category of volatile nutrients present in the medium. Their obvious decreasing trend in the cancer cell lines (see supporting Figure S2) is consistent with the known increase in nutrient uptake and consumption for the fast proliferation and low efficiency of energy usage of cancer cells. They provide indirect evidence for differences in cell proliferation and metabolism between normal and cancerous cells.
The opposite trend was observed for the compounds found in negative ion mode. Fatty acids are necessary for energy storage, membrane proliferation and the generation of signaling molecules19. Because cancer cells have a disorder in cell growth and proliferation, they need to accumulate more metabolic intermediates for cellular building blocks; cancer cells often exhibit a shift toward fatty acid synthesis. The peak at m/z 85.0295 was assigned to but-2-enoic acid (see http://www.hmdb.ca/), which is involved in fatty acid biosynthesis. Its concentration was found to be higher (median ratio = 1.43, p < 0.05) in cancer cells than in normal cells. Moreover, it consistently showed higher intensity than that in the normal cell line, with the highest concentration in the T47D and the lowest in the SKBR-3 cell lines. Considering the cell proliferation rate is just opposite to the concentration change trend, this suggests that the molecule with m/z 85.0295 and some of other smaller fatty acids may act as the intermediates for the biosynthesis of longer chain fatty acids in rapidly proliferating cancer cells.
Besides fast proliferation, cancer cells often show extensive generation of reactive oxygen species (ROS) and an altered redox status. The generated ROS can react with the polyunsaturated fatty acids of lipid membranes and induce lipid peroxidation20. The major end product of lipid peroxidation is 4-hydroxynonenal (4-HNE), which is the tentative assignment for the peak at m/z 183.1389, which is also among the 9 molecules that showed significant differences in negative ion mode (Table 1).
It was reported that increased aldehyde levels can be detected in the exhaled breath of lung cancer patients, which may be due to lipid peroxidation and related to 4-HNE, the major end product21. Here we consistently observed an increased level of 4-HNE in the headspace of culture medium for breast cancer cell lines. It has been reported that through exposure to different pathologies that involve the production of ROS, lipid peroxidation resulting in 4-HNE production increases and the local concentration of 4-HNE in tissues can increase to 10 μM or more22.
Moreover, the peak of 4-HNE was also the highest in T47D cells and the lowest for the SKBR-3 cells. Its level in all cancer cell lines was consistently higher than that in the normal cell line (see supporting Figure S1). It has been reported that a moderate increase in ROS can promote cell proliferation and differentiation23,24, whereas excessive amounts of ROS may cause oxidative damage and then inhibit tumor cell growth19. Several research reports indicate that a 4-HNE up-regulation in cancer cells induces a reduction of cell proliferation and an increase of apoptosis25,26. Moreover, it was reported that by analyzing a time-course series of samples, some dynamic network biomarkers that are able to distinguish a pre-disease state from normal and disease states could be identified27. Concentrations of 4-HNE and other discriminating volatile fatty acids varied for different stages of breast cancer, i.e., they may relate to the occurrence and development of cancer. Therefore, we hypothesize that T47D showed a low proliferation rate partly because of the high concentration of 4-HNE, while a low concentration of 4-HNE in SKBR-3 may contribute to the fast proliferation.
In conclusion, when analyzing the metabolic signature of human breast cancer cells (MDA-MB-231) versus normal human mammary cells (HMLE), we were able to find volatile compounds in the headspace of conditioned culture medium that showed different concentrations between cancer and normal cells. Statistical analysis of the volatile signature revealed a clear distinction between cancer cells and normal cells. In addition, from high-resolution mass spectrometry, we could tentatively assign the chemical composition of some of the most discriminating molecules and formulate hypotheses about their biological values. Thus, we conclude that cancerous cells can release a characteristic volatile signature. In particular, some small fatty acids are produced in higher concentrations compared with normal cells. Complementarily to routine metabolomics studies aimed at disease biomarker discovery, these volatile fatty acids can be easily detected and quantified in vivo in exhaled human breath28,29, a much more readily accessible sample, in real time. Further work will be needed to assess whether these compounds may ultimately be exhaled in abnormally high concentrations by cancer patients. If so, these volatile fatty acids can potentially be developed for use as disease biomarkers to serve as a basis for a noninvasive and easy-to-use diagnostic tool21.
Methods
Sample preparation
The T47D, SKBR-3, MDA-MB-231 (breast cancer) and HMLE (normal mammary) cell lines (all from ATCC) were plated using 2 × 106 cells in 10-cm cell culture plates the day before analysis. The cells were washed twice with PBS to remove the growth medium, which was then replaced with DMEM (Gibco). The DMEM was incubated with the cell cultures or alone at +37°C for 24 hours. The DMEM was then collected, centrifuged to remove possible cell contamination and stored at −80°C until analysis. Ten biological replicates for each cell type were prepared for analysis to check the reproducibility.
Real-time secondary electrospray ionization mass spectrometric measurements
A quadrupole time-of-flight (QTOF; Waters, Synapt G2) was slightly modified to measure the headspace of culture medium. As previously described30, a 3 mL aliquot of the supernatant of culture medium was put in a 50 mL-flask with two necks in a water bath at 40°C. Air was bubbled through the liquid via the neck of the flask and a Teflon tube, at a flow rate of 5 L/min. In this fashion, the headspace of the culture medium was transported to the orifice of the mass spectrometer via a Teflon tube (50 cm long, 3 mm i.d.), which was connected to the curtain gas channel of the mass spectrometer. This sampling tube was coated with heating tape maintained at a temperature of 40°C. To make sure that the same amount of headspace was delivered to the mass spectrometer in every analysis, the bubbling air flow was maintained for 6 seconds in every measurement (time for ~4 scans of the mass spectrometer). The headspace mingled with an electrospray plume formed by a lab-built nano electrospray source (PicoTip emitter, i.d. 20 μm), where some molecules get ionized and subsequently are detected by the mass analyzer. This ionization technique is dubbed secondary electrospray ionization (SESI)13,14,15,17,18,31,32,33,34,35,36 and has already been applied for real-time analysis of trace gaseous analytes in different applications (e.g. human skin and breath vapors28,29,37,38,39,40,41,42,43,44,45 and bacterial volatiles46,47,48,49,50. The electrosprayed solution (0.2% aqueous formic acid) was infused at ~100 nL/min (2 kV, ~700 nA). The electrospray tip was located ~6 mm from the sampling cone on the symmetry axis.
Data analysis
Five consecutive headspace mass spectra of each sample were averaged and the same number of scans of blank (background) spectra in between measurements were subtracted, using Water's MassLynx software. The processed raw mass spectra were then saved as .txt files. These were post processed using commercial software (MATLAB, R2012b, Mathworks Inc., USA). First, each mass spectrum was interpolated to 150,000 mass-to-charge (m/z) values (55–350 Da, in steps of 0.00197 Da). After centroiding, the 150,000 m/z values were reduced to 4,794 features in negative ion mode and 4,957 in positive mode. For each polarity, the 100 (5 technical replicates × 20 biological replicates) mass spectra were normalized using the linear normalization method51. Finally, we assembled a 100 × 4,794 (negative mode) and 100 × 4,957 (positive mode) matrices with each of these normalized mass spectra. Each of the 100 mass spectra was categorized either as normal (n = 50) or cancerous (n = 50).
Next, the mean values of all five technical replicates were calculated, resulting in a 20 × 4,794 data matrix in negative ion mode and a 20 × 4,975 data matrix in positive ion mode. The m/z values with a median intensity of less than 300 counts in each group were discarded as noise.
We then sought to reduce the dimensionality of the matrices by applying a filtering approach52,53. A widely-used filter method in bioinformatics is to apply a univariate criterion separately on each feature. In our case, to identify the individual peaks that showed significant differences between the normal cells and breast cancer cells, we conducted a Mann–Whitney U test and kept the peaks that yielded p-values < 0.05 for further analysis. Further feature subset reduction was accomplished by dropping the isotopic peaks and signals that, despite background subtraction, were not clearly (as assessed by visual inspection) above the background level. As a result, a total of 16 m/z values (9 in negative ion mode and 7 in positive ion mode) were retained for further analysis.
Thus, the original 20 × 4,794 and 20 × 4,975 matrices were reduced to 20 × 9 and 20 × 7, respectively. These positive and negative ion mode matrices were merged into a single 20 × 16 matrix, which was then subjected to principal component analysis (PCA).
The first two principal components explaining most of the variance were identified and used for class prediction. Finally, the cell samples' categories (i.e. cancer vs. normal) were predicted using a k-nearest neighbor algorithm (k = 1; Euclidean distance) in a leave-one-out cross validation. Thus, each sample was out of the model once and assigned to the class of its nearest neighbor. The process was then repeated for all the samples and the overall classification performance was assessed.
References
Jemal, A. et al. Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 (2009).
Denkert, C. et al. Metabolomics of human breast cancer: New approaches for tumor typing and biomarker discovery. Genome Medicine 4, 37–46 (2012).
Odunsi, K. et al. Detection of peithelial ovarian cancer using 1H-NMR-based metabonomics. Int. J. Cancer 113, 782–788 (2005).
Asiago, V. et al. Early Detection of recurrent breast cancer using metabolite profiling. Cancer Res. 70, 8309–8318 (2010).
Griffin, J. L. & Shockcor, J. P. Metabolic profiles of cancer cells. Nat. Rev. 4, 551–561 (2004).
Catherine, O. Uncovering the metabolomic fingerprint of breast cancer. Int. J. Biochem Cell Biol. 43, 1010–1020 (2011).
Williams, H. & Pembroke, A. Sniffer dogs in the melanoma clinic. Lancet 1, 734 (1989).
Church, J. & Williams, H. Another sniffer dog for the clinic. Lancet. 358, 930 (2001).
McCulloch, M. et al. Diagnostic accuracy of canine scent detection in early- and late-stage lung and breast cancers. Integr. Cancer Ther. 5, 1–10 (2006).
Filipiak, W. et al. TD-GC-MS Analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer Epidemiol. Biomarkers Prev. 19, 182–195 (2010).
Smith, D., Wang, T., Sulé-Suso, J., Spanel, P. & El Haj, A. Quantification of acetaldehyde released by lung cancer cells in vitro using selected ion flow tube mass spectrometry. Rapid Commun. Mass Spectrom. 17, 845–850 (2003).
Bullinger, D. et al. Metabolic signature of breast cancer cell line MCF-7: Profiling of modified nucleosides via LC-IT MS coupling. BMC Biochem. 8, 25–39 (2007).
Wu, C., Siems, W. F. & Hill, H. H. Secondary electrospray ionization ion mobility spectrometry/mass spectrometry of illicit drugs. Anal. Chem. 72, 396–403 (2000).
Chen, Y. H., Hill, H. H. & Wittmer, D. P. Analytical merit of electrospray ion mobility spectrometry as a chromatographic detector. J. Microcolumn Sep. 6, 515–24 (1994).
Kiselev, P. & Fenn, J. B. ESIMS Analysis of Vapors at Trace Levels. Proceedings of the 49th ASMS Conference on Mass Spectrometry and Allied Topics, Chicago, Illinois, 2001 May 27–31.
Fuerstenau, S., Kiselev, P. & Fenn, J. B. ESIMS in the Analysis of Trace Species in Gases. Proceedings of the 47th ASMS Conference on Mass Spectrometry, Dallas (TX), 1999.
Fuerstenau, S. Aggregation and Fragmentation in an electrospray ion source. New Haven (CT): Yale University, 1994.
Whitehouse, C. M., Levin, F. & Meng, C. K. Further Adventures With An Electrospray Ion Source. Proceedings of the 34th ASMS Conference, Cincinnati, OH, 1986.
Currie, E., Schulze, A., Zechner, R., Walther, T. C. & Farese, R. V. Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 18, http://dx.doi.org/10.1016/j.cmet.2013.05.017 (2013).
Barrera, G. Oxidative stress and lipid peroxidation products in cancer progression and therapy. ISRN Oncol. 2012, 137289 (2012).
Hakim, M. et al. Volatile organic compounds of lung cancer and possible biochemical pathways. Chem. Rev. 112, 5949–5966 (2012).
Forman, H. J. Reactive oxygen species and α, β-unsaturated aldehydes as second messengers in signal transduction. Ann. N. Y. Acad. Sci. 1203, 35–44 (2010).
Boonstra, J. & Post, J. A. Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene 337, 1–13 (2004).
Schafer, F. Q. & Buettner, G. R. Redox environment of the cell as viewed through the tedox state of the glutathione disulfide/glutathione couple. Free Radical Biol. Med. 30, 1191–1212 (2001).
Fazio, V. M. et al. 4-Hydroxynonenal, a product of cellular lipid peroxidation, which modulates c-myc and globin gene expression in K562 erythroleukemic cells. Cancer Res. 52, 4866–4871 (1992).
Barrera, G. et al. Inhibition of c-myc expression induced by 4-hydroxynonenal, a product of lipid peroxidation, in the HL-60 human leukemic cell line. Biochem. Biophys. Res. Commun. 203, 553–561 (1994).
Liu, R., Wang, X., Aihara, K. & Chen, L. Early diagnosis of complex diseases by molecular biomarkers, network biomarkers and dynamical network biomarkers. Med. res. rev. 10.1002/med.21293 (2013).
Martinez-Lozano Sinues, P. & Fernández de la Mora, J. Direct analysis of fatty acid vapors in breath by electrospray ionization and atmospheric pressure ionization-mass spectrometry. Anal. Chem. 80, 8210–8215 (2008).
Martinez-Lozano Sinues, P., Zingaro, L., Finiguerra, A. & Cristoni, S. Secondary electrospray ionization-mass spectrometry: Breath study on a control group. J. Breath Res. 5, 016002 (2011).
Martinez-Lozano Sinues, P. et al. Mass spectrometry fingerprinting coupled to national institute of standards and technology mass spectral search algorithm for pattern recognition. Anal. Chim. Acta. 755, 28–36 (2012).
Fernandez de la Mora, J. Ionization of vapor molecules by an electrospray cloud. Int. J. Mass Spectrom. 300, 182–93 (2011).
Vidal-de-Miguel, G., Macía, M., Pinacho, P. & Blanco, J. Low-Sample Flow Secondary Electrospray Ionization: Improving Vapor Ionization Efficiency. Anal. Chem. 84, 8475–9 (2012).
Vidal-de-Miguel, G. & Herrero, A. Secondary Electro Spray Ionization of complex vapor mixtures. Theoretical and experimental approach. J. Am. Soc. Mass Spectrom. 23, 1085–1096 (2012).
Martinez-Lozano Sinues, P., Criado, E. & Vidal, G. Mechanistic study on the ionization of trace gases by an electrospray plume. Int. J. Mass Spectrom. 313, 21–29 (2012).
Martinez-Lozano Sinues, P., Rus, J., Fernández de la Mora, G., Hernández, M. & Fernández de la Mora, J. Secondary Electrospray Ionization (SESI) of Ambient Vapors for Explosive Detection at Concentrations Below Parts Per Trillion. J. Am. Soc. Mass Spectrom. 20, 287–294 (2009).
Dillon, L. A. et al. Optimisation of secondary electrospray ionisation (SESI) for the trace determination of gas-phase volatile organic compounds. Analyst 135, 306–314 (2010).
Martinez-Lozano Sinues, P. et al. Breath Analysis in Real Time by Mass Spectrometry in Chronic Obstructive Pulmonary Disease. Respiration. 87, 301–310 (2014).
Martinez-Lozano Sinues, P., Kohler, M. & Zenobi, R. Monitoring diurnal changes in exhaled human breath. Anal. Chem. 85, 369–373 (2013).
Martinez-Lozano Sinues, P., Kohler, M. & Zenobi, R. Human breath analysis may support the existence of individual metabolic phenotypes. PLoS ONE. 8, e59909 (2013).
Martinez-Lozano Sinues, P., Zenobi, R. & Kohler, M. Analysis of the exhalome: A diagnostic tool of the future. CHEST Journal. 144, 746–749 (2013).
Martinez-Lozano Sinues, P. Mass spectrometric study of cutaneous volatiles by secondary electrospray ionization. Int. J. Mass Spectrom. 282, 128–132 (2009).
Martinez-Lozano Sinues, P. & Fernández de la Mora, J. On-line Detection of Human Skin Vapors. J. Am. Soc. Mass Spectrom. 20, 1060–1063 (2009).
Martinez-Lozano Sinues, P. & de la Mora, J. F. Electrospray ionization of volatiles in breath. Int. J. Mass Spectrom. 265, 68–72 (2007).
Reynolds, J. C. et al. Detection of volatile organic compounds in breath using thermal desorption electrospray ionization-ion mobility-mass spectrometry. Anal Chem. 82, 2139–2144 (2010).
Berchtold, C., Meier, L., Steinhoff, R. & Zenobi, R. A new strategy based on real-time secondary electrospray ionization and high-resolution mass spectrometry to discriminate endogenous and exogenous compounds in exhaled breath. Metabolomics. 1–11 (2013).
Zhu, J., Bean, H. D., Wargo, M., Leclair, L. & Hill, J. E. Detecting bacterial lung infections: in vivo evaluation of in vitro volatile fingerprints. J. Breath Res. 7, 016003 (2013).
Zhu, J., Bean, H. D., Jiménez-Díaz, J. & Hill, J. E. Secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting of multiple bacterial lung pathogens, a mouse model study. Journal of Applied Physiology 114, 1544–1549 (2013).
Zhu, J. et al. Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: from initial infection to clearance. J. Breath Res. 7, 037106 (2013).
Bean, H. D., Zhu, J. & Hill, J. E. Characterizing Bacterial Volatiles using Secondary Electrospray Ionization Mass Spectrometry (SESI-MS). J. Vis. Exp. e2664 (2011).
Zhu, J., Bean, H. D., Kuo, Y. M. & Hill, J. E. Fast Detection of Volatile Organic Compounds from Bacterial Cultures by Secondary Electrospray Ionization-Mass Spectrometry. J. Clin. Microbiol. 48, 4426–4431 (2010).
Borgaonkar, S. P., Hocker, H., Shin, H. & Markey, M. K. Comparison of normalization methods for the identification of biomarkers using MALDI-TOF and SELDI-TOF mass spectra. OMICS 1, 115–126 (2010).
Das, S. Filters, Wrappers and a Boosting-Based Hybrid for Feature Selection. ICML '01: Proceedings of the Eighteenth International Conference on Machine Learning. Morgan Kaufmann Publishers Inc. 74–81 (2001).
Kohavi, R. & John, G. H. Wrappers for feature subset selection. Artificial Intelligence. 97, 273–324 (1997).
Acknowledgements
JJH was supported by the National Instrumentation Program (2011YQ170067) and the China Scholarship Council. JJH thanks her Ph.D. supervisor, Prof. Zeper Abliz (Peking Union Medical College) for supporting her Ph.D. exchange program. MH was supported by the Sigrid Jusélius Foundation and the Instrumentarium Foundation. This research was supported by a Marie Curie European Reintegration Grant (PMLS) within the 7th European Community Framework Programme (276860).
Author information
Authors and Affiliations
Contributions
R.Z., P.M.L.S. and J.J.H. designed the research; M.H. prepared the culture medium for cell lines and measured the proliferation rate of cell lines; J.J.H., P.M.L.S. and X.L. performed MS measurement of the volatiles; P.M.L.S. and J.J.H. analyzed data; M.D. supervised the culture medium preparation and proliferation rate measurement; J.J.H. and P.M.L.S. wrote the manuscript; all authors contributed to editing the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplemental Information
Supplementary Information
Suppl. Fig. 3
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
He, J., Sinues, PL., Hollmén, M. et al. Fingerprinting Breast Cancer vs. Normal Mammary Cells by Mass Spectrometric Analysis of Volatiles. Sci Rep 4, 5196 (2014). https://doi.org/10.1038/srep05196
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05196
This article is cited by
-
Investigation of VOCs associated with different characteristics of breast cancer cells
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.