Abstract
Mitochondria share a common ancestor with the Alphaproteobacteria, but determining their precise origins is challenging due to inherent difficulties in phylogenetically reconstructing ancient evolutionary events. Nonetheless, phylogenetic accuracy improves with more refined tools and expanded taxon sampling. We investigated mitochondrial origins with the benefit of new, deeply branching genome sequences from the ancient and prolific SAR11 clade of Alphaproteobacteria and publicly available alphaproteobacterial and mitochondrial genome sequences. Using the automated phylogenomic pipeline Hal, we systematically studied the effect of taxon sampling and missing data to accommodate small mitochondrial genomes. The evidence supports a common origin of mitochondria and SAR11 as a sister group to the Rickettsiales. The simplest explanation of these data is that mitochondria evolved from a planktonic marine alphaproteobacterial lineage that participated in multiple inter-specific cell colonization events, in some cases yielding parasitic relationships, but in at least one case producing a symbiosis that characterizes modern eukaryotic life.
Similar content being viewed by others
Introduction
Vital to the evolution of all known eukaryotic cells was the endosymbiotic event that resulted in the permanent acquisition of bacteria that through time were transformed into mitochondria. Even eukaryotes that were previously thought to lack mitochondria have now been shown to contain some remnants of that original endosymbiosis, either through the maintenance of hydrogenosomes, mitosomes, or nuclear genes of mitochondrial origin1,2. Although argument continues about the circumstances of the founding endosymbiotic event2,3, current hypotheses, making use of mitochondrial genome and proteome data, agree that the mitochondrial ancestor was most closely related to Alphaproteobacteria4,5,6,7,8,9,10,11. The rooting of the ancestral mitochondria within the Alphaproteobacteria has not yet been resolved, but better phylogenetic methodology and new mitochondrial and bacterial genome sequences offer opportunities to refine understanding of this singular evolutionary event.
The sequencing of the first Rickettsia genome provided phylogenomic evidence for the previously established theory that the mitochondrial root was within the Rickettsiales5,10,12,13 (and references therein). This theory was intellectually satisfying because all known Rickettsia have extremely reduced genomes and obligate intracellular lifestyles, although there was evidence to suggest that the genome reduction in mitochondria and Rickettsia probably happened independently5,10,11. While the rooting of mitochondria within the Rickettsiales has been debated based on other phylogenomic studies4,7,14, it has gained support more recently with the addition of more genome sequences from taxa in the Rickettsiales, including the globally important SAR11 clade6,15.
As free-living, predominantly marine bacteria, the phylogenetically diverse16 organisms of the ubiquitous SAR11 clade are unique in the Rickettsiales order. Like other Rickettsiales, SAR11 have small genomes and, consequently, limited metabolic versatility5,7,17,18,19. However, SAR11 genomes bear distinctly different evolutionary signatures from other Rickettsiales genomes, indicating that selection for a streamlined genome, rather than genetic drift, has been the path of genome reduction in these organisms9,17,19. For example, the sequence of Candidatus Pelagibacter ubique HTCC1062 has few recent gene duplications, phage, transposons, or pseudogenes, has extremely small intergenic regions and is optimized for a highly efficient lifestyle17, whereas organisms like Rickettsia and those of the Anaplasmataceae contain large numbers of duplications, putatively inactive genes, larger intergenic regions, and/or mobile elements, indicative of the different selective pressures of obligate intracellular life7,18,20,21. Despite of the differences in evolutionary history and lifestyle, there is phylogenomic evidence for a shared common ancestor between SAR11 organisms and the Rickettsiales and for rooting of the mitochondria in this group6, but the latter is still a matter of debate22. Here, we systematically tested the phylogenomic relationship of the SAR11 clade to the mitochondria using an automated pipeline and with the benefit of new sequences from deeply branching SAR11 genomes23.
Results
Experimental design and logic
We framed our phylogenomic testing around the following logic. First, we analyzed a subset (127) of available alphaproteobacterial genomes, including seven SAR11 genomes, with six outgroup species from the Gamma-, Beta- and Deltaproteobacteria (Table S1, Fig. 1a). These outgroup taxa had a range of G+C content (40–67%, mean ∼55%). Second, we tested the SAR11 genomes with the Rickettsiales separately for comparison to see if there was an observable effect of removing high G+C content taxa, using a smaller “median” outgroup with a mean G+C content of ∼53% (Table S2, Fig. 1b). Third, with these established topologies, we then introduced mitochondrial sequences (Table S3) and evaluated the stability of the topology to variations in taxon sampling, gap-removal strategy and allowed missing data using the same outgroup (Fig. 2). Fourth, we examined the consistency of these topologies to G+C bias of the outgroup taxa at low (mean ∼40%) and high (mean ∼67%) values (Figs. 4, 5).
Summary of phylogenomic analyses without mitochondrial sequences.
The consensus trees for both the Alphaproteobacteria (a) and the Rickettsiales (b) across 10–60% allowed missing data are shown with concomitant information regarding number of OCs and amino acid sites for each individual tree. Rows are allowed missing data (10–60%), columns are gap-removal strategy (C- conservative, L- liberal, R- remgaps). Branching frequencies are noted at the nodes. # - this tree is featured in Fig. 6. Other abbreviations: Ana- Anaplasmataceae, Rick- Rickettsiaceae, Rhodosp- Rhodospirillales, Sph- Sphingomonadales, CHP- Caulobacterales + Hyphomonadaceae + Parvularculales, Rhod- Rhodobacteraceae.
Summary of phylogenomic analyses including mitochondrial sequences.
Data is divided by each dataset and corresponding tree topology (a–e). Internal tables for each dataset show the relevant information for each individual tree output. Rows are allowed missing data (10–60%), columns are gap-removal strategy (C- conservative, L- liberal, R- remgaps). Bootstrap support values ≥ 80% are reported, grey circles indicate a value between 79 and 51%, open circles ≤ 50%, Inc- incongruence in non-mitochondrial branch order compared with trees built without mitochondrial sequences and other studies. † - HIMB59 grouped with the mitochondria. * - two mitochondrial sequences grouped with the Ehrlichia. ‡ - mitochondria grouped with the SAR11 clade. # - this tree is featured in Fig. 7. Other abbreviations: mt- mitochondria, Ana- Anaplasmataceae, Rick- Rickettsiaceae, Rhodosp- Rhodospirillales, Sph- Sphingomonadales, Ca- Caulobacterales, Pa- Parvularculales, OG- outgroup, BS- bootstrap, OCs- orthologous clusters.
Alphaproteobacteria tree topologies determined with outgroups biased by low (a) and high (b) G+C content.
Each observed tree is shown with an associated table for the corresponding missing data percentage(s) (rows) and gap-removal parameter(s) (columns: C- conservative, L- liberal, R- remgaps). Overall included orthologous clusters (OCs) and amino acid sites for each dataset are specified at the bottom. * - bootstrap support for that node < 60%. OG- outgroup, Ana- Anaplasmataceae, Rick- Rickettsiaceae, Rhodosp- Rhodospirillales, Sph- Sphingomonadales, CHP- Caulobacterales, Hyphomonadaceae, Parvularculales, Rhod- Rhodobacteraceae.
Summary of phylogenomic analyses with mitochondrial sequences using biased outgroups.
Data is divided by each dataset and corresponding tree topology (a–f). Internal tables for each dataset show the relevant information for each individual tree output, grouped according to G+C bias of the outgroup taxa. Rows are allowed missing data (40 and 60%), columns are gap-removal strategy (C- conservative, L- liberal, R- remgaps). Bootstrap support values ≥ 80% are reported, grey circles indicate a value between 79 and 51%, open circles ≤ 50%, Inc- incongruence in non-mitochondrial branch order compared with trees built without mitochondrial sequences and other studies. † - HIMB59 grouped with the mitochondria. ‡ - mitochondria grouped with the SAR11 clade. Other abbreviations: mt- mitochondria, Ana- Anaplasmataceae, Rick- Rickettsiaceae, Rhodosp- Rhodospirillales, Sph- Sphingomonadales, Ca- Caulobacterales, Pa- Parvularculales, OG- outgroup, BS- bootstrap, OCs- orthologous clusters.
Within the Hal pipeline, the cutoff percentage for the minimum number of taxa required for an orthologous cluster (OC) to be included (missing data) can be specified. The deleterious effect of missing data on phylogenetic analysis can be offset by the advantageous effect of additional taxon sampling if there are a large number of total sites and can improve accuracy24,25. Thus, taxa with fewer protein coding genes, such as mitochondria, can be incorporated into a robust phylogenetic analysis that makes use of the signal generated by larger amounts of sequence from bacterial taxa with higher gene content. We analyzed each initial dataset over the range of 10–60% allowed missing data at 10% increments to assess the stability of tree topology across differing numbers of total included OCs and to allow for the inclusion of taxa with varying amounts of coding sequence. Biased outgroup tests were then done on a subset of these missing data values (40 and 60%).
The variations in taxon sampling, gap-removal strategy and allowed missing data were put in place specifically to address potential systematic error known to effect phylogenomic analyses26,27 (See Discussion). Changing these parameters causes different OCs, as well as different numbers of OCs, to be used for each analysis. This strategy can reveal error resulting from gene-specific heterotachy; likewise, altering the sampling of both ingroup and outgroup taxa with aberrant G+C content can reveal perturbations of branch order caused by compositional bias. In addition to these parameter variations, we also used the CAT model of rate heterogeneity, for which a Bayesian implementation has been shown to be helpful for overcoming both compositional signal error and long-branch attraction brought on by rate signal error26,28,29,30.
Phylogenomic analysis of the Alphaproteobacteria and Rickettsiales
The majority of trees generated for the 127 Alphaproteobacteria genomes over 10–60% allowed missing data with different gap-removal stringencies supported predicted family- and order-level groupings, with the previously established exception that the Caulobacterales, Hyphomonadaceae and Parvularculales always grouped together, thus making the Rhodobacterales polyphyletic6 (Fig. 1a). Two of the 18 trees failed to reconstruct the Rickettsiales as a monophyletic group and two separated the Rhodospirillales, although all four had poor bootstrap support at these nodes (Fig. S1, SI tree files). Sixteen of eighteen trees formed the characteristic bifurcation of the Rickettsiales separate from the remaining orders of the phylum, which has been demonstrated previously by different methods6,31 (Fig. 1a). The branching order of clades between the Rhodospirillales and the Rhizobiales was unstable (Fig. S1); however, the consensus tree demonstrates that this was usually limited to the positioning of the Rhodobacteraceae and the Sphingomonadales (Fig. 1a). Regardless, the Rickettsiales consistently grouped away from the remainder of the tree and, in accordance with previous studies6,32, the SAR11 clade was monophyletic with the Rickettsiaceae and the Anaplasmataceae in 16 of 18 trees; 2 of these 16 grouped SAR11 with the Rickettsiaceae instead of basal to the Rickettsiaceae and the Anaplasmataceae. The two trees where the Rickettsiales was paraphyletic had poorly supported nodes for the Rickettsiaceae/SAR11 group and the Anaplasmataceae (Fig. S1). A separate analysis adding the sequence of Magnetococcus sp. MC-1, for which a potential position within the Alphaproteobacteria has been suggested8, placed this sequence among the outgroup taxa in all trees at 50 and 20% allowed missing data (data not shown).
We further focused our analysis on the Rickettsiales alone to verify the topology revealed by the complete alphaproteobacterial dataset with more included OCs and all 44 available genomes for the order and to more firmly establish the relationship of the SAR11 clade to the Rickettsiales without the compositional signal of other, higher G+C content organisms. Gamma- and Betaproteobacteria sequences from the previous analysis served as outgroups (Table S2). The tree topology was consistent across 10–60% missing data, which included from 130 to 349 OCs (Fig. 1b). Importantly, at the family and genus level, this consensus tree agreed with the topology of the Rickettsiales order that was observed in the complete alphaproteobacterial phylogeny.
Inclusion of mitochondrial genome sequences
To test the phylogenomic placement of the mitochondria and SAR11 clades among the Alphaproteobacteria and monitor the stability of the resulting topologies to sampling perturbation, we adopted the following strategy: six different datasets with variation in sampling among mitochondrial or alphaproteobacterial taxa (Table S2) were tested across the range of 10–60% included missing data (Fig. 2). Established orders of Alphaproteobacteria were added in increments, using available genome sequences for each order and the same outgroups from the Rickettsiales analysis. We chose mitochondrial genome sequences (Table S3) as a function of the phylogenetic affiliation of their eukaryotic hosts. We included mitochondria from the Excavata, Chromalveolata and Archaeplastida, as these clades have mitochondrial members considered “ancestral”33,1,34 and are phylogenetically distinct from the “Unikonts,” which include Metazoans35,36, a group with highly variable mitochondrial evolutionary rates37,38. Further, in order to maximize the number of included OCs, we restricted our analyses to those mitochondria within these groups that contained more than 30 protein-coding genes (Table S3).
Figure 2 summarizes the results from the five datasets with a majority of trees that had branching patterns consistent among all datasets considered in this study. A sixth dataset, which included the Rhodobacterales, was in almost all cases incongruent in non-mitochondrial topology and/or had very poor bootstrap support at the major nodes or the mitochondrial node (SI tree files). The results from this dataset were therefore not included. Total OCs, the number of aligned sites and bootstrap values for the mitochondrial node are reported in the figure. Trees that were incongruent with monophyletic relationships established without the mitochondrial sequences are noted as "Inc”; these topologies were usually poorly supported at major nodes where the branching order differed (SI tree files). 18 mitochondrial sequences (Table S3a) were tested with increasing numbers of alphaproteobacterial sequences (Fig. 2a, c–e) and a group of 48 mitochondrial sequences (Table S3b) was tested with just the Rickettsiales, including SAR11 (Fig. 2b). Two alternative topologies were observed with the largest dataset that was consistent with the monophyletic relationships we had established (Fig. 2e). In general, with the exception of the dataset in Fig. 2c, in all trees and all datasets with the median outgroup, the SAR11 were monophyletic with the mitochondria, or else were the outgroup to a clade joining mitochondria, the Anaplasmataceae and the Rickettsiaceae, in either case linking them closely to mitochondrial origins (Fig. 2a, b, d, e).
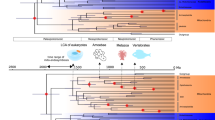
When Rickettsiales were the only additional sequences in the tree, support for the monophyly of mitochondria and the SAR11 clade was very high and consistent across variation in missing data, gap-removal strategy and the variation in taxon sampling within the mitochondria (Fig. 2a, b). One tree showed an anomalous pattern in which two mitochondrial sequences grouped with the Ehrlichia sequences (Fig. 2b asterisk) and two trees were incongruent with the trees that did not include mitochondria (Fig. 2b- “Inc”). Addition of the Rhodospirillales sequences changed the branching pattern and caused the mitochondria to group with the Rickettsiaceae instead of the SAR11 clade in all but one tree (Fig. 2c). In this dataset and those in Fig. 2d–e, trees with the fewest OCs were incongruent. However, inclusion of missing data resulted in considerably more signal and consistency in topology across datasets. Additionally, inclusion of more taxa from the orders Sphingomonadales, Caulobacterales and Parvularculales reversed the topological change in Fig. 2c and again grouped the mitochondria as either monophyletic with SAR11 (Fig. 2d, e) or in a paraphyletic relationship with SAR11 as the outgroup to a clade joining mitochondria, the Anaplasmataceae and the Rickettsiaceae (Fig. 2e). More trees placed the mitochondria and SAR11 as monophyletic (7) than as paraphyletic (5) and were better supported. In several trees, the deepest branching SAR11 strain, HIMB59, grouped with the mitochondria (Figs. 2, 7 daggers), thus making the monophyly of these groups more pronounced.
Example tree of mitochondrial taxa included with the Rickettsiales, Rhodospirillales and Sphingomonadales using the median outgroup ( Fig. 2d ) showing all taxa, 50% missing data included, liberal gap removal.
Dagger indicates the single taxon instability of HIMB59, observed in several trees throughout the datasets in Fig. 2. G+C % is indicated for each taxon, numbers below 50% are in bold. Scale bar indicates 0.4 changes per position. Bootstrap values are indicated at nodes.
The addition of more taxa did not have a significant effect on the total numbers of OCs used in the analyses, except in the case where additional mitochondria sequences were used (Fig. 2b). While the relative contribution of each taxon decreased as the allowed missing data parameter was increased and more OCs were included, the absolute number of OCs increased for most taxa, thereby improving the net signal contributed by a given taxon. Figure 3 shows this effect for the mitochondrial taxa in Fig. 2d in trees with liberal gap removal (column L), where an increase in OCs was observed for every taxon; in some the increase was 70% or greater across the tested range.
Mitochondrial OCs identified in the 18 mitochondria phylogeny alignments as a function of included missing data and total identified OCs for the liberal gap-removal trees in Fig. 2d .
Key: Bn, Brassica napus mt; Ca, Chlorokybus atmophyticus mt; Cg, Chaetosphaeridium globosum mt; Cm, Cyanidioschyzon merolae mt; Cv, Chara vulgaris mt; Ha, Hemiselmis andersenii mt; Mj, Malawimonas jakobiformis mt; Mp, Marchantia polymorpha mt; Mv, Mesostigma viride mt; Ng, Naegleria gruberi mt; No, Nephroselmis olivacea mt; Ov, Ostreococcus tauri mt; Pa, Pseudendoclonium akinetum mt; Pl, Pylaiella littoralis mt; Pp, Porphyra purpurea mt; Ps, Phytophthora soja mt; Ra, Reclinomonas americana mt; Rs, Rhodomonas salina mt.
Effects of outgroup G+C content bias
The deep-branching position of the Rickettsiales and SAR11 make them particularly vulnerable to compositional bias-based attraction artifacts depending on the outgroup taxa. The initial outgroup selection was the same as that of Williams et al.6 (less two taxa) and covered a range of G+C content (40–67%) which we utilized specifically to avoid such artifacts. However, since the monophyly of SAR11 and the Rickettsiales is essential to examining the placement of the mitochondria in that group we tested the Alphaproteobacteria dataset in the presence of biased outgroups of both low and high G+C content. We also tested the stability of the datasets in Fig. 2 with biased outgoups. The number and taxonomic affiliation at the class level, of outgroup taxa was kept constant to avoid significantly changing the number of included OCs or introducing additional artifacts based on branch distances. The biased Alphaproteobacteria outgroups spanned G+C ranges of 32–46%, mean ∼40% and 65–75%, mean ∼67%, for the “low” and “high” groups, respectively (Table S1). The biased outgroups for the datasets including mitochondria were chosen with similar means: 41% (low) and 66% (high) (Table S2). All datasets were tested with 40 and 60% included missing data levels, as these values provided very robust signal and provided a large difference in OCs (more than 2× for most datsets). All gap-removal strategies were also tested.
The main effect of biased outgroups on the Alphaproteobacteria was to alter the topological relationships of the major clades established using the median outgroup. However, the major clades remained intact, with the exception of two trees using the low G+C biased outgroup (Fig. 4). These placed SAR11 outside of the Rickettsiales, although this topology had much weaker bootstrap support than trees which placed SAR11 as monophyletic with the Rickettsiales (Fig. 4a, SI tree files) and SAR11 was still placed in the Rickettsiales by the majority of trees. Trees with the high G+C biased outgroup no longer placed the Rickettsiales as the basal group to the tree and instead pulled higher G+C content clades towards the outgroup as would be expected from such biasing. (Fig. 4b, SI tree files). While this bias disrupted the general placement of the Rickettsiales as the deepest branch of the Alphaproteobacteria, SAR11 still formed a monophyletic group with the Rickettsiales.
For the analyses which included mitochondrial sequences and varying numbers of alphaproteobacterial orders (Fig. 5), the results were mostly similar to those using the median outgroup, with the exception of the dataset in Fig. 5d, which did not agree with Fig. 2d. Including both high and low G+C bias, 75% of the trees grouped mitochondria with the Rickettsiaceae, instead of SAR11 as was shown with the median outgroup. Three of the trees did still group SAR11 and mitochondria together as in Fig. 2d. Fig. 5b low outgroup trees with 40% missing data were incongruent (“Inc”) to the whole Alphaproteobacteria because they placed the Anaplasmataceae as basal to the Rickettsiaceae/SAR11/mitochondria group, but still supported the monophyly of SAR11 and mitochondria (SI tree files). The largest dataset including the Rickettsiales, Rhodospirillales, Sphingomonadales, Caulobacterales and Parvularculales with the mitochondria again showed two separate tree topologies, distributed across both biases and again those which supported the monophyly of SAR11 and mitochondria were better supported (Fig. 5e). Thus, the datasets in Fig. 5a,b, c and e generally agreed with the median outgroup analyses shown in Fig. 2 regardless of the outgroup bias.
Discussion
We systematically tested phylogenomic relationships of SAR11 to Alphaproteobacteria and mitochondria using new, deeply branching SAR11 genomes we sequenced and over 60 additional genomes that were not available to previous studies6. The automated pipeline Hal allowed for methodic consistency across 216 separate phylogenomic analyses. There were some limitations to the pipeline: at low levels of allowed missing data, it found very few acceptable clusters across all taxa (data not shown) and there was an upper limit to the number of taxa that could be included with consistent results if the genomes were substantially different. Nevertheless, by combining the results of a variety of sampling and analysis perturbations, we have compiled a consistent view of the SAR11 clade within the Alphaproteobacteria and the relationship of mitochondria to this clade.
The observed position of SAR11 within the Rickettsiales and the Alphaproteobacteria agreed with a previous phylogenomic analysis of the Alphaproteobacteria6, as well as other phylogenomic and 16S rRNA gene taxonomy studies18,31,32,39. Also, the consensus topology we observed for the entire Alphaproteobacteria supported the findings of Williams et al.6 (Figs. 1a, S1). When taxon sampling was limited to the Rickettsiales, the observed relationships agreed with those of the complete Alphaproteobacteria tree (Fig. 1b) and all trees, regardless of gap-removal strategy, confirmed a Rickettsia phylogeny by Gillespie et al. created from 731 core genes18 (SI tree files). Importantly, the use of biased outgroups also supported the placement of SAR11 in a monophyletic relationship with the Rickettsiales (Fig. 4). Thus, SAR11 placement within the Rickettsiales was mostly stable across perturbations of missing data, gap-removal strategy, taxon sampling and outgroup G+C bias, making this topology the most likely given the currently available genomes. Further, based on comparisons of branch lengths between the SAR11 clade, Rickettsiaceae and the Anaplasmataceae across the various trees (see an example in Figs. 6 and 7) and the 80% 16S rRNA gene sequence identity between HIMB59 (Group V) and HTCC1062 (Group I), SAR11 comprises enough phylogenetic diversity to be considered a distinct family, proposed here as Pelagibacteraceae, fam. nov.
When mitochondrial sequences were included in the analysis, the relationship of SAR11 to the Rickettsiales did not change and most trees supported the monophyly of SAR11 and the mitochondria (Figs. 2, 5). The trees in Fig. 2c and Fig. 5c, d demonstrate the need for alteration of taxon sampling (see below) and attention to outgroup G+C bias when testing for phylogenetic signal. Although many of the trees from these two datasets provided support for the alternative monophyletic relationship of SAR11 and the Rickettsiaceae, they did not agree with the overall majority of trees. The dataset in Fig. 5d was not wholly consistent with itself, with only 25% of the trees in agreement when the same dataset was used without a biased outgroup (Fig. 2d). Also, the incongruence between Fig. 5d and Fig. 2d demonstrated that this selection of taxa was particularly sensitive to outgroup G+C bias. However, the remaining datasets were in good agreement regardless of outgroup G+C content (Figs. 2, 5). Overall, the analyses more frequently placed the mitochondria as monophyletic with SAR11 (85/128 trees), than either between SAR11, the Rickettsiaceae and the Anaplasmataceae (11/128 trees), or monophyletic with the Rickettsiaceae (31/128 trees).
We chose our approach to test the robustness of tree topology to perturbations and thereby minimize the effects of horizontal gene transfer (HGT) and systematic error. The consistency of topologies and the high average bootstrap values across datasets, as well as the large number of included OCs in most of the trees means that the effect of HGT was probably overcome with increasing amounts of sequence signal38. However, systematic error cannot be overcome by such means. While phylogenomic analysis can dramatically reduce stochastic error, the increase in characters does not relieve the problems of systematic errors, which remain constant26,27,40,30. Using the terminology of Philippe et al.26, systematic errors (non-phylogenetic signal) can be broken into three categories: rate signal, compositional signal and heterotachous signal (for detailed reviews see Philippe et al.26 and Delsuc et al.27). Briefly, rate signal arises from variation in the evolutionary rates of different taxa, which can lead to long-branch attraction (LBA) artifacts of highly divergent taxa. Compositional signal is the result of G+C content bias between organisms in the analysis and can result in organisms of similar G+C content being artificially grouped together. Heterotachous signal comes from site and/or gene-specific rate variation within and between organisms.
All of these signal biases were addressed in our analysis. We made use of the CAT model of rate heterogeneity in RAxML specifically to deal with LBA29 and created separate amino acid substitution models for each OC that was included in the tree-building process. Alteration of allowed missing data, which changed both the number and the type of OCs used throughout the analysis, was expected to limit the effects of heterotachous signal. Another potential problem for the phylogenetic placement of mitochondria and SAR11 was that in general, Rickettsiales, including SAR11, have much lower G+C content than other Alphaproteobacteria (Fig. 6). In our analyses, organisms with low G+C content were scattered throughout the alphaproteobacterial tree and did not consistently branch together (Fig. 6), even in the face of substantial outgroup bias (Figs. 4, 5, SI tree files). Also, the monophyly of the mitochondria across datasets is noteworthy considering the wide G+C range among mitochondria, especially considering the presence of the Anaplasma, which have a much higher G+C content than the other Rickettsiales (Fig. 7). Analysis of the mitochondrial sequences with the Rickettsiales alone addressed both LBA and compositional signal error by removing higher G+C content organisms and those with shorter branches. The observation that biased outgroups and the addition of higher G+C Alphaproteobacteria sequences generally did not disrupt the topology, across a variety of included orthologous clusters, provides further support for the monophyly of SAR11 and mitochondria.
While the various techniques employed here are helpful to uncovering true phylogenetic signal, taxon sampling is perhaps the most important countermeasure to systematic errors26,27,30.The effect of taxon sampling was clearly shown by the aberrant trees in Fig. 2c, which had high bootstrap support but were incongruent with the remaining trees. We were able to demonstrate consistency across large sampling differences, both in taxa and included OCs. The effect of allowing more missing data was to increase the number of OCs used in both the overall analyses as well as per taxa, which resulted in phylogenies that were more robust, in agreement with previous work24,25 (and references therein). This had significant practical impact on our study because inclusion of missing data also allowed the inclusion of taxa with less coding sequence - specifically, mitochondria.
Yet, mitochondrial genome size is the result of massive gene transfer from the original endosymbiont to the nuclear genome and therefore the signal which we made use of for this study has to be viewed as a subset of available information. Many of the original proto-mitochondrion genes retain enough phylogenetic signal to identify them as having come from ancestral Alphaproteobacteria, in spite of the fact that Eukaryotic substitution of analogous genes has occurred13,41,42 (and references within). An initial study of these nuclear encoded mitochondrial genes showed close relationships to organisms throughout the Alphproteobacteria22. In light of the results we present here, close phylogenetic scrutiny of nuclear encoded genes that originated from mitochondria, in the context of new Alphaproteobacteria sequences such as SAR11, would appear to be a promising direction for future research on this topic. Also, obtaining sequences from deeper branching Alphaproteobacteria and examination of these relationships with alternative methods, such as supertrees, or sequence-independent features like gene order, rare genomic changes and other technologies currently being developed and tested (see Delsuc et al.27 for a review), could provide additional perspective on the evolutionary links between SAR11, mitochondria, Rickettsiales and other Alphaproteobacteria.
While the variety of issues discussed above warrant caution, our findings nevertheless provide support for an intriguing new hypothesis: that mitochondria evolved from free living ancestors of the SAR11 clade, rather than from a lineage of intracellular organisms, as previously suggested10,12. Prior work indicated that the ancestral mitochondrion was most likely an aerobic heterotroph2,41 with respiratory genes closely related to those in Rickettsiaceae (reviewed in10). The physiology of SAR11 organisms is consistent with the proposed proto-mitochondria: all strains are heterotrophic, obligate aerobes, with predicted aerobic electron transport chains like those in mitochondria, composed of NADH dehydrogenases, fumarate reductases, ubiquinones, cytochrome bc1 complexes, cytochrome c, cytochrome c oxidases and ATP synthases. Several of these genes were among the orthologous clusters used to construct the phylogenies, along with ribosomal proteins (e.g., Table S4) and thus support the close evolutionary relationship between these pathways/genes in SAR11 organisms and the mitochondria.
Modern SAR11 cells have small genomes imposed by selection for efficient replication in nutrient limited systems17, which have evolved unusual nutritional requirements that would pre-adapt them to closer dependence on other organisms43,44,45. While many explanations have been offered for the singular origin of mitochondria2 (and references therein), the findings we present support a simple but plausible scenario: mitochondria originated in the aerobic ocean, involving a lineage that repeatedly colonized other species and ultimately gave rise to the largest free living bacterial populations known. Contemporary marine systems contain abundant Archaea and Eukarya, both theorized hosts for the original mitochondrial symbiosis2. We cannot know if the current conditions are similar to those at the time of the mitochondrial and SAR11 ancestor, but we can add to the scientific discussion by postulating that it was likely a free-living marine organism and focus interest broadly on the mitochondria, the SAR11 clade and Rickettsiales to identify shared properties that led to their successful divergence.
Methods
We recently sequenced five additional SAR11 genomes from three phylogenetically distinct clades, corresponding to the subgroups Ia, III and a new, distantly related subgroup (Group V)46 (Figs. 6 & 7), which were annotated consistently with others used in this study by IMG47 (http://img.jgi.doe.gov/cgi-bin/m/main.cgi). We made exclusive use of publicly available bacterial genome sequences from the IMG database to control for gene-calling methodological differences across databases. The 127 taxa used in the whole alphaproteobacterial dataset (Table S1) were a subset of the total available sequences chosen to represent all sequenced genera. Given that some species are highly overrepresented in the database, we omitted several taxa from these overrepresented groups. Datasets with mitochondrial sequences included most available genome sequences from the respective Alphaproteobacteria orders (Table S2). Mitochondrial sequences (Table S3) were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). The sequences were analyzed using Hal, an automated pipeline for phylogenetic analysis of genomic protein sequence data.
The Hal pipeline23,48 (http://aftol.org/pages/Halweb3.htm; http://sourceforge.net/projects/bio-hal/) consists of a set of Perl scripts that automates a series of phylogenomic analyses using existing software and sequence analysis programs and was executed on a 64-bit Linux cluster operating Red Hat Linux 3.2.3, Linux version 2.4.21. For this study, the Hal pipeline directed the following analyses: Protein sequences were imported in fasta format from sequenced genomes listed in Table S1–3 and subjected to an all-vs-all BLASTP with the output E-values provided to the program MCL49. Using a Markov Clustering algorithm, MCL grouped proteins into orthologous clusters (OCs) as function of the all-vs-all BLASTP E-values. Clustering was executed across a range of stringencies (inflation parameters 1.1 – 5.0) and OCs were filtered for any redundant clusters (clusters found at more than one inflation parameter), clusters containing more than one protein per genome (multi-copy OC) and clusters containing proteins whose best reciprocal BLAST hit was outside of the cluster (clusters more likely to contain paralogs). Clusters were also filtered across a range of missing values settings (10–60%) whereby a cluster may contain only one protein per genome but a defined percentage of the genomes may be missing from the cluster. This function allows for the incorporation for more single-copy clusters when such proteins are either missing from a given genome or missing from a genome annotation.
Protein sequences for accepted OCs were extracted from their respective genome fasta sequence files and aligned using MUSCLE50 with default settings. To accommodate for problematic regions of the alignments, three separate alignments were created for each protein alignment: one removing all gap-containing columns (remgaps) and two removing problematic regions of the alignments based on the default conservative (Glocks-con) and liberal (Gblocks-lib) options of the program GBlocks51. Best models of amino acid substitution for each protein alignment were estimated using ProtTest52. Individual protein alignments were concatenated into one super-alignment and analyzed using RAxML53 with the PROTCAT setting for the rate model and with each protein partition of the super-alignment assigned its best model of amino acid substitution. Nodal support was estimated based on 100 bootstrap replications using the rapid bootstrapping option as implemented in RAxML. For each analysis, three phylogenetic trees were generated, representing the three super-alignments (remgaps, GBlocks-con and GBlocks-lib) produced as part of the automated alignment routine.
References
Gray, M. W., Lang, B. F. & Burge, G. Mitochondria of Protists. Annu. Rev. Genet. 38, 477–524 (2004).
Embley, T. M. & Martin, W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006).
Koonin, E. V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11, 209–221 (2010).
Fitzpatrick, D. A., Creevey, C. J. & McInerney, J. O. Genome Phylogenies Indicate a Meaningful α-Proteobacterial Phylogeny and Support a Grouping of the Mitochondria with the Rickettsiales . Mol. Biol. Evol. 23, 74–85 (2005).
Andersson, S. G. E. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–143 (1998).
Williams, K. P., Sobral, B. W. & Dickerman, A. W. A Robust Species Tree for the Alphaproteobacteria . J. Bacteriol. 189, 4578–4586 (2007).
Wu, M. et al. Phylogenomics of the Reproductive Parasite Wolbachia pipientisw Mel: A Streamlined Genome Overrun by Mobile Genetic Elements. PLoS Biol. 2, 0327–0341 (2004).
Esser, C., Martin, W. & Dagan, T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol. Letters 3, 180–184 (2007).
Ettema, T. J. G. & Andersson, S. G. E. The α-proteobacteria: the Darwin finches of the bacterial world. Biol. Letters 5, 429–432 (2009).
Andersson, S. G. E., Karlberg, O., Canback, B. & Kurland, C. On the origin of mitochondria: a genomics perspective. Philos. T. R. Soc. B. 358, 165–179 (2003).
Gray, M. W., Burger, G. & Lang, B. F. The origin and early evolution of mitochondria. Genome Biol. 2, 1–5 (2001).
Emelyanov, V. V. Mitochondrial connection to the origin of the eukaryotic cell. Eur J Biochem 270, 1599–1618 (2003).
Kurland, C. G. & Andersson, S. G. E. Origin and Evolution of the Mitochondrial Proteome. Microbiol. Mol. Biol. Rev. 64, 786–820 (2000).
Esser, C. et al. A Genome Phylogeny for Mitochondria Among α-Proteobacteria and a Predominantly Eubacterial Ancestry of Yeast Nuclear Genes. Mol. Biol. Evol. 21, 1643–1660 (2004).
Morris, R. et al. SAR 11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002).
Stingl, U., Tripp, H. J. & Giovannoni, S. J. Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 1, 361–371 (2007).
Giovannoni, S. J. et al. Genome Streamlining in a Cosmopolitan Oceanic Bacterium. Science 309, 1242–1245 (2005).
Gillespie, J. J. et al. Rickettsia Phylogenomics: Unwinding the Intricacies of Obligate Intracellular Life. PLoS ONE 3, 1–34 (2008).
Wilhelm, L., Tripp, H. J., Givan, S., Smith, D. P. & Giovannoni, S. J. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct 2, (2007).
Brayton, K. A. et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Nat. Acad. Sci. U.S.A. 102, 844–849 (2005).
Collins, N. E. et al. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Nat. Acad. Sci. U.S.A. 102, 838–843 (2005).
Atteia, A. et al. A Proteomic Survey of Chlamydomonas reinhardtii Mitochondria Sheds New Light on the Metabolic Plasticity of the Organelle and on the Nature of the α-Proteobacterial Mitochondrial Ancestor. Mol. Biol. Evol. 26, 1533–1548 (2009).
Robbertse, B., Yoder, R. J., Boyd, A., Reeves, J. B. & Spatafora, J. W. Hal: an Automated Pipeline for Phylogenetic Analyses of Genomic Data. PLoS Curr. Tree of Life 4, (2011).
Philippe, H. et al. Phylogenomics of Eukaryotes: Impact of Missing Data on Large Alignments. Mol Biol Evol 21, 1740–1752 (2004).
Wiens, J. J. Missing data and the design of phylogenetic analyses. J. Biomed. Inform. 39, 34–42 (2006).
Philippe, H., Delsuc, F., Brinkmann, H. & Lartillot, N. PHYLOGENOMICS. Annu. Rev. Ecol. Evol. Syst. 36, 541–562 (2005).
Delsuc, F., Brinkmann, H. & Philippe, H. Phylogenomics and the Reconstruction of the Tree of Life. Nat Rev Genet 6, 361–375 (2005).
Lartillot, N. & Philippe, H. A Bayesian Mixture Model for Across-Site Heterogeneities in the Amino-Acid Replacement Process. Mol. Biol. Evol. 21, 1095–1109 (2004).
Lartillot, N., Brinkmann, H. & Philippe, H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7, 1–14 (2007).
Philippe, H. et al. Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough. PLoS Biol 9, e1000602 (2011).
Lee, K. The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Micr. 55, 1907–1919 (2005).
Logares, R., Brate, J., Heinrich, F., Shalchian-Tabrizi, K. & Bertilsson, S. Infrequent Transitions between Saline and Fresh Waters in One of the Most Abundant Microbial Lineages (SAR11). Mol. Biol. Evol. 27, 347–357 (2010).
Gray, M. W., Burger, G. & Lang, B. F. Mitochondrial Evolution. Science 283, 1476–1481 (1999).
Lang, B. R. et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387, 493–497 (1997).
Hampl, V. et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. PNAS 106, 3859–3864 (2009).
Burki, F., Shalchian-Tabrizi, K. & Pawlowski, J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol. Letters 4, 366–369 (2008).
Brown, W. M., George, M., Jr. & Wilson, A. C. Rapid evolution of animal mitochondrial DNA. Proc. Nat. Acad. Sci. U.S.A. 76, 1967–1971 (1979).
Shearer, T. L., Oppen, M. V., Romano, S. & Worheide, G. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol. Ecol. 11, 2475–2487 (2002).
Rappe, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002).
Jeffroy, O., Brinkmann, H., Delsuc, F. & Philippe, H. Phylogenomics: the beginning of incongruence? Trends Genet. 22, 225–231 (2006).
Gabaldon, T. & Huynen, M. A. Reconstruction of the Proto-Mitochondrial Metabolism. Science 301, 609 (2003).
Gross, J. & Bhattacharya, D. Mitochondrial and plastid evolution in eukaryotes: an outsiders' perspective. Nat. Rev. Genet. 10, 495–505 (2009).
Tripp, H. J. et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744 (2008).
Tripp, H. J. et al. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ. Microbiol. 11, 230–238 (2009).
Schwalbach, M. S., Tripp, H. J., Steindler, L., Smith, D. P. & Giovannoni, S. J. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ. Microbiol. 12, 490–500 (2010).
Moeseneder, M. M., Arrieta, J. M. & Herndl, G. J. A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol. Ecol. 51, 341–352 (2005).
Markowitz, V. M. et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res 36, D534–D538 (2008).
Robbertse, B., Reeves, J. B., Schoch, C. L. & Spatafora, J. W. A phylogenomic analysis of the Ascomycota. Fungal Gent. and Biol. 43, 715–725 (2006).
van Dongen, S. Graph Clustering by Flow Simulation. (2000).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004).
Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Acknowledgements
We thank Christopher M. Sullivan and the Oregon State University Center for Genome Research and Biocomputing (CGRB) for management of and troubleshooting for the bioinformatics infrastructure. Sequencing, assembly and annotation of SAR11 genomes was supported by the Gordon and Betty Moore Foundation Marine Microbiology Initiative, as part of its Marine Microbial Sequencing Project (http://www.moore.org/marinemicro), by an individual investigator award to SJG and a grant from the National Science Foundation Science and Technology Center, Center for Microbial Oceanography: Research and Education(Award No. EF0424599)to MSR. This material is based upon work supported by the National Science Foundation under Award No. DBI-1003269 to JCT.
Author information
Authors and Affiliations
Contributions
AB assisted with bioinformatics analysis, program writing and created the SI website, MJH, JG and PC cultivated cells, extracted DNA for genome sequencing and aided in annotation, MSR provided three SAR11 genome sequences and assisted in study design, RJY wrote the Hal scripts and assisted with bioinformatics analysis, BR oversaw Hal development and implementation, JWS oversaw Hal development and implementation and assisted in study design, JCT and SJG designed the study, SJG and JWS assisted in writing the paper, JCT wrote the paper/created the figures, compiled the data and performed the bioinformatics analysis. All authors discussed the results and commented on the paper.
Ethics declarations
Competing interests
All tree files, super alignments and model files used in the study are available at: http://sar11-phylogenomics.cgrb.oregonstate.edu.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Thrash, J., Boyd, A., Huggett, M. et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 1, 13 (2011). https://doi.org/10.1038/srep00013
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00013
This article is cited by
-
Genomic profiling and characteristics of a C1 degrading heterotrophic fresh-water bacterium Paracoccus sp. strain DMF
Archives of Microbiology (2024)
-
Ecophysiology and genomics of the brackish water adapted SAR11 subclade IIIa
The ISME Journal (2023)
-
Influence of short and long term processes on SAR11 communities in open ocean and coastal systems
ISME Communications (2022)
-
Wolbachia: endosymbiont of onchocercid nematodes and their vectors
Parasites & Vectors (2021)
-
Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria
Nature Ecology & Evolution (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.