« Prev Next »
Eukaryotic genomes are replicated with almost absolute fidelity every time cells divide. However, conditions for DNA replication are rarely ideal. For example, environmental toxins or drugs cause DNA damage and interfere with DNA replication. How does a cell circumvent such problems? How does a cell generate an exact copy of genetic materials under such conditions?
During every cell division, a cell must duplicate its chromosomal DNA through a process called DNA replication. The duplicated DNA is then segregated into two "daughter" cells that inherit the same genetic information. This process is called chromosome segregation. Because DNA is a depository of genetic information, DNA replication and segregation must be achieved with extreme fidelity. Failure of these processes can cause mutations and chromosome rearrangements, leading to diseases or even death (Abraham 2001; Kastan & Bartek 2004).
Healthy cells can perform DNA replication with almost absolute accuracy most of the time. Considering that a eukaryotic cell contains millions or billions of DNA base pairs, this is a remarkable accomplishment. However, the conditions for DNA synthesis are rarely ideal, with several obstacles challenging the DNA replication machinery (Branzei & Foiani 2007; Heller & Marians 2006; Lambert, Froget & Carr 2007). It seems that DNA replication is far more difficult than one might think. So, what could interfere with DNA replication?
Obstacles on Template DNA Generate Stalled Replication Forks
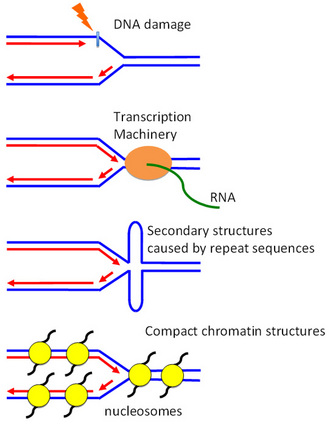
Replication blockage is not only caused by DNA damage (Figure 1). Proteins bound on DNA also provide significant obstacles for DNA replication, as replication fork proteins can collide with them and become displaced from DNA. These include various DNA binding proteins such as the transcription machinery.
Another physical barrier could be the DNA structure itself. Repeated DNA sequences form complex 3-dimensional structures such as telomeres, centromeres and ribosomal DNA and may be difficult to replicate. Highly ordered chromatin structures also cause replication problems, as they are very compact and wrap around histone proteins, and in turn result in highly ordered structures. All of these can cause replication fork stalling (Figure 1).
One way to envision the replication fork is to think about bicycling. It is quite easy to keep your balance when you are moving forward. But once you stop, it is difficult to keep upright without stepping on the ground. If you don't step on the ground, you may fall down. Similarly, stalled forks are inherently unstable and prone to collapse, posing a serious threat to genomic integrity because stalled forks can break or rearrange (Figure 2) (Branzei & Foiani 2007; Heller & Marians 2006; Lambert, Froget, & Carr 2007). How does a cell circumvent these problems? How does a cell achieve accurate replication of genomes under such daunting conditions?
The DNA Replication Checkpoint Arrests the Cell Cycle
In 1989, Leland Hartwell and Ted Weinert proposed that a cell possesses dedicated quality control systems that monitor problems on DNA templates. These systems are referred to as cell cycle checkpoints and are regulated by a number of proteins (Figure 3) (Hartwell & Weinert 1989). The checkpoints detect various issues found on DNA. Once checkpoint proteins identify these issues, the cell activates signal transduction pathways in order to arrest the progression of the cell cycle and allow adequate time to fix the problems on DNA. We also know that the checkpoints activate DNA repair programs to coordinate with cell cycle arrest (Branzei & Foiani 2008; Paulsen & Cimprich 2007; Aguilera & Gomez-Gonzalez 2008).
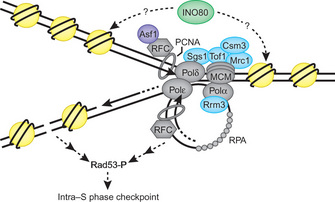
We know much about how the replication checkpoint fixes the damage by coordinating cell cycle arrest and DNA repair programs (Figure 2) (Branzei & Foiani 2008; Paulsen & Cimprich 2007; Aguilera & Gomez-Gonzalez 2008). When the fork stalls or breaks, the cell sequentially activates signal transduction proteins to ensure that offspring inherit accurate copies of parental DNA. Atop this system stand the ATM/ATR related protein kinases, which transduce the signal to arrest the cell cycle. ATR and ATM phosphorylate downstream kinases such as Chk1 and Chk2 are important to inactivate the cell cycle engine, CDK1, by inhibiting Cdc25 and activating Wee1 (Figure 2) (Carr 2002; Nyberg et al. 2002; Boddy & Russell 2001). These activities prevent the cell from moving into the later phases of the cell cycle, ensuring that the cells do not pass on incomplete or altered genetic information to the daughter cells. ATR and ATM also phosphorylate histone H2AX and other DNA repair proteins to promote efficient repair of DNA damage (Durocher & Jackson 2001). Therefore, checkpoint kinases couple DNA repair with cell cycle arrest in order to fix the problems on DNA (Figure 2). What else, then, does the replication checkpoint do?
Protecting DNA Replication Forks
Preserving genomic integrity at the unwound, exposed DNA when the fork stalls is another major responsibility of the replication checkpoint (Branzei & Foiani 2008; Paulsen & Cimprich 2007; Aguilera & Gomez-Gonzalez 2008). Therefore, it is also important to prevent DNA damage at the replication fork by maintaining assembly of the replisome (the replication machinery) components and DNA structures in replication competent states when forks stall. Recent studies identified a group of proteins that are required to stabilize replication forks (Figure 3). These fork protectors may include proteins related to Timeless, Tipin, and Claspin (Abraham 2001; Katou et al. 2003; Leman et al. 2010; McFarlane, Mian & Dalgaard 2010; Noguchi et al. 2004). While we are still trying to understand how these and other proteins stabilize replication forks (McFarlane, Mian & Dalgaard 2010; Branzei & Foiani 2010), it seems that these fork protectors have at least the following functions (Figure 3):
- Coordinating various activities required for DNA synthesis. For example, it is important to couple DNA replication activity of DNA polymerases with the unwinding activity of DNA helicases at the replication fork.
- Stabilizing replisome components at the replication fork when the fork stalls, so that the fork can re-start after the problems are solved.
- Preventing fork breakage. If the fork is broken, fork protectors may be required to re-assemble replisome components. This function is critical to resume DNA replication once DNA is repaired.
- Stabilizing unusual DNA structures at the stalled fork and hence avoiding unwanted DNA recombination.
- Protecting the replication forks in a configuration that is recognized by replication checkpoint proteins (Figure 2).
Multiple Processes at the Replication Fork: Beyond DNA Replication
Several concurrent processes are occurring during DNA replication at the replication fork, so fork protectors may have other functions. For example, sister chromatid cohesion is an essential process required for accurate segregation of chromosomes. After DNA replication, the cohesion mechanism holds duplicated sister chromatids together until they are ready for separation at mitosis. Importantly, there are cohesins — proteins that are required for cohesion — that are loaded onto the chromosome before DNA replication. Although exactly how it happens is controversial, scientists think cohesin proteins form ring-like structures and wrap around the chromosome. Therefore, cells establish sister chromatid cohesion at the replication fork when the fork passes through the cohesin ring during DNA replication (Nasmyth & Haering 2009). Indeed, downregulation of fork protectors results in cohesion defects (Leman et al. 2010; McFarlane, Mian & Dalgaard 2010).
Another example of cellular processes at the replication fork is chromatin regulation. Chromatin contains histone proteins that are required for the assembly of nucleosomes, the fundamental unit of chromatin. Chromatin structure has a large impact on DNA replication and repair programs. During DNA replication, cells need to disassemble nucleosomes ahead of the replication fork and reassemble them behind the fork (Figure 5). At the same time, in S-phase, cells need to synthesize new histone proteins which will also be deposited to newly replicated DNA. Such orchestration of DNA replication processes in the context of chromatin — so called DNA replication-coupled nucleosome assembly — is extremely important for accurate transmission of genetic and epigenetic information (Annunziato 2005; Groth et al. 2007; Henikoff, Furuyama & Ahmad 2004). Therefore, it is likely that replication fork proteins are involved in regulation of chromatin structures (Figure 4).
Summary
When the fork stalls, the replication checkpoint steps in and identifies the problems with the DNA. This checkpoint activates a signal transduction cascade to arrest the cell cycle and coordinate with the DNA repair programs. This checkpoint also directly protects replication forks in order to prevent genetic instability. Thus, it is obvious that the replication fork is a major site of cellular regulation that ultimately preserves genomic integrity. The fork is not only for DNA replication. As described above, multiple events take place at the fork. Currently, we are trying to understand how multiple processes are regulated and coordinated at the replication fork. Future studies will show us how regulation of DNA replication relates to other cellular processes at the replication fork.
References and Recommended Reading
Abraham, R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15, 2177–2196 (2001).
Aguilera, A. & Gomez-Gonzalez, B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9, 204–217 (2008).
Annunziato, A. T. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 280, 12065–12068 (2005).
Boddy, M. N. & Russell, P. DNA replication checkpoint. Curr. Biol. 11, R953–R956 (2001).
Branzei, D. & Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 6, 994–1003 (2007).
Branzei, D. & Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9, 297–308 (2008).
Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11, 208–219 (2010).
Carr, A. M. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amst) 1, 983–994 (2002).
Durocher, D. & Jackson, S. P. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol 13, 225–231 (2001).
Groth, A. et al. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Hartwell, L. H. & Weinert, T. A. Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634 (1989).
Heller, R. C. & Marians, K. J. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7, 932–943 (2006).
Henikoff, S., Furuyama, T. & Ahmad, K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet 20, 320–326 (2004).
Kastan, M. B. & Bartek, J. Cell-cycle checkpoints and cancer. Nature 432, 316–323 (2004).
Katou, Y. et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003).
Lambert, S., Froget, B. & Carr, A. M. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 6, 1042–1061 (2007).
Leman, A. R. et al. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci 123, 660–670 (2010).
McFarlane, R. J., Mian, S. & Dalgaard, J. Z. The many facets of the Tim-Tipin protein families' roles in chromosome biology. Cell Cycle 9, 700–705 (2010).
Nasmyth, K. & Haering, C. H. Cohesin: its roles and mechanisms. Annu Rev Genet 43, 525–558 (2009).
Noguchi, E. et al. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24, 8342–8355 (2004).
Nyberg, K. A.et al. TOWARD MAINTAINING THE GENOME: DNA Damage and Replication Checkpoints. Annu Rev Genet 36, 617–656 (2002).
Paulsen, R. D. & Cimprich, K. A. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 6, 953–966 (2007).
Uhlmann, F. A matter of choice: the establishment of sister chromatid cohesion. EMBO reports 10, 1095-1102 (2009).




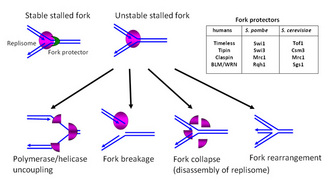
 Figure 3: Fork protection
Figure 3: Fork protection


























