« Prev Next »
All mammals have basically the same set of genes, yet there are obviously some significant differences that distinguish the various species. Researchers currently think that much of mammalian morphological diversity involves cis regulatory regions, or stretches of DNA outside the actual coding region of a gene that are responsible for switching the gene on and off.
However, an important paper by Fondon and Garner (2004) suggests that there is yet another source of variation: tandem repeats. Tandem repeats are short lengths of DNA that are repeated multiple times within a gene, anywhere from a handful of times to more than a hundred. These sequences are also called VNTRs, or variable number tandem repeats, because different individuals within a population may have different numbers of repeats. VNTRs are relatively easy to detect with molecular tools, and researchers know that populations (humans included) may carry a large reservoir of different numbers of repeats. For example, one person might carry three tandem repeats in a particular gene, while another person might bear 15, with no obvious differences between the two individuals that can be traced to that particular gene. So, the question is, what, if anything, does having a different number of tandem repeats do to an organism?
The Basic Premise of Fondon and Garner's Study
In their 2004 study, researchers John Fondon and Harold Garner set out to answer this question by first looking for populations that exhibited large and obvious morphological differences between individuals, and then looking within these individuals' genomes to see whether those differences could be correlated with the number of tandem repeats present. The duo decided to use domestic dogs as their population; after all, not only are dogs diverse, but dog breeders are notoriously picky about shape and character, and purebred dogs have been under intense selection for specific attributes for many years.
Once a range of morphologies in a particular trait, such as snout shape, has been identified, one can ask whether this range is reflected in the number of repeats in any genes. Thus, Fondon and Garner examined 142 dogs from 92 different breeds, and they looked at 37 different tandem repeats in 17 genes in each dog. The genes selected were those that encoded transcription factors that were at least suspected of playing a role in the formation of specific morphologies during development. Fifteen of the 17 genes turned out to have multiple alleles that varied in the number of copies of repeats they contained.
Tandem Repeats and Mutations

The fact that Fondon and Garner found a substantial amount of genetic variation in tandem repeat number is not at all surprising, because tandem repeats are subject to very high mutation rates. This increased probability of mutation (up to 100,000 times greater than the probability of a point mutation) exists because tandem repeats are prone to a kind of error called slipped-strand mispairing. Tandem repeats contain many copies of the same short sequence over and over, so it is easy for the two strands of DNA to get misaligned in this local region—the GTAC sequence on one strand could base-pair with the first CATG in the other strand, or the second, or the third, for example. If the strands are mispaired, then DNA replicating enzymes can err and either clip off some of the repeats or add extra repeats (Figure 1). This represents a special kind of error in that the DNA changes do not occur in random nucleotides, and they produce only different numbers of repeats. Also, note that this lack of fidelity in copying tandem repeats means that such repeats are only found in regions of genes that can tolerate some variability.
Interestingly, slipped-strand mispairing can be foiled by point mutations, even to synonymous codons, within a tandem repeat. Here, a small change in the DNA sequence gives the replication machinery a local difference that can be used to properly align the two strands. Over time, however, a stable tandem repeat will accumulate these small changes and lose its repeated character. On the other hand, a deletion caused by slipped-strand mispairing can remove a point difference, and subsequent mispairing can then expand the sequence, producing a repeat free of imperfections. Thus, one measure of how much selection for variation has occurred within a tandem repeat is its purity. If there are few interruptions in the perfection of the repeat, there has been much deletion and expansion going on within the sequence throughout its history. Conversely, if there are multiple deviations from perfect repetition, then the sequence has not undergone much length variation in the recent past.
Repeat Variants in Dogs and Their Association with Morphology
The purity of a tandem repeat sequence is therefore a measure of how much selection for new variants has occurred in an organism's lineage. Based on this principle, Fondon and Garner compared the same repeat loci in humans and dogs, and they found that the dog repeats were purer than the human repeats in 29 of 36 cases; in the other seven cases, the dog and human repeats were of equal purity. This finding strongly suggests that the variations in dogs are not just random, neutral changes, but rather, they are the outcome of recent selection at these loci.
Thus, Fondon and Garner determined that there are multiple interesting gene variants in dogs, and these variants have apparently undergone selection. But what effect do the repeats have? Let's consider two specific genes—Runx-2 and Alx-4—as examples.
Runx-2
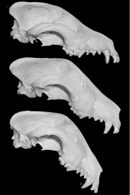
The Runx-2 (runt-related transcription factor 2) gene is related to the Drosophila pair-rule gene runt, which is involved in segmentation. In vertebrates, one of the functions of Runx-2 is to regulate the differentiation of osteoblasts, which are the cells responsible for forming new bone. Runx-2 contains two repeats, one coding for 18–20 glutamines (the poly-Q region) and another coding for 12–17 alanines (the poly-A region). A statistical comparison of the total length of both of these repeats (poly-Q + poly-A) with various parameters of canine skull size revealed a correlation with the dogs' midface length, as well as with a property called clinorhynchy, or dorsoventral nose bend. To better understand what clinorhynchy looks like, think about the distinctive, long nose of bull terriers, which features a downward droop (Figure 2). Bull terriers tend to have a short pair of tandem repeats in Runx-2, and they have long midfaces and pronounced downturn of the snout. The breed has been intentionally selected for this trait, and museum specimens over the past 70 years show increased prominence of this feature (Figure 3).
In reality, the relationship between Runx-2 and canine morphology is not as simple as short repeat length equals downturned snout. Remember, one of the ways that the activity of transcription factors is regulated is by binding, and chains of amino acids can affect how transcription factors interact and bind with one another. Moreover, it turns out that polyglutamine can increase the rate of transcription in the genes that it regulates, while polyalanine can reduce it. Of course, the Runx-2 protein has both a polyglutamine (poly-Q) and a polyalanine (poly-A) chain. Thus, what might matter more in a situation such as this (where two competing components modulate activity) is the ratio of poly-Q to poly-A. Indeed, this ratio shows an even stronger correlation with clinorhynchy than does the total combined length of the poly-Q and poly-A regions (Figure 4).
Alx-4
A second gene example is Alx-4 (aristaless-like homeobox 4). This gene is also related to a transcription factor found in Drosophila, and knocking out the gene in mice produces individuals with six toes. In Fondon and Garner's study, one specific allele of this gene, Alx-4Δ51, was found in only one breed of dog, the Great Pyrenees. One peculiarity of this breed is hind limb polydactyly—purebreds are supposed to have a double dewclaw, for a total of six digits on each of their hind legs. Thus, it is not surprising that the Alx-4Δ51 allele features a deletion that knocks out 51 nucleotides from a specific tandem repeat, for a total loss of 17 amino acids. All Great Pyrenees with polydactyly have this particular deletion (Figure 5); moreover, in Fondon and Garner's study, the only Great Pyrenees that did not have the extra dewclaw carried the full-length tandem repeat.
The Benefits and Limitations of an Extreme Example
The good news about these findings is that they demonstrate another mode by which morphological diversity can be added to a population relatively rapidly, as well as another mechanism for fine-tuning evolution. Because tandem repeats are common in the vertebrate genome, these repeats could clearly be a reservoir of variation and a robust and flexible way to add new variations to populations.
There are some limitations to Fondon and Garner's results, though. First, this study focused on an extreme case: purebred dogs that have undergone very strong selection for specific and, in some cases, outright deleterious characteristics. Thus, we simply don't know how important this mode of evolutionary change is under less-artificial conditions. Second, this study and others like it have revealed only correlations, not experimental perturbations. While these correlations are convincing, at some point in the future, it would be helpful to see direct manipulation of the poly-Q/poly-A ratio in the Runx-2 gene of a collie, for instance, to give it the downturned nose of a bull terrier. Finally, it would also be beneficial to obtain additional correlative evidence through developmental studies of the patterns of Runx-2 and Alx-4 gene expression in dog embryos to see exactly how these variations play out.

References and Recommended Reading
Fondon, J. W., & Garner, H. R. Molecular origins of rapid and continuous morphological evolution. Proceedings of the National Academy of Sciences 101, 18058–18063 (2004)
Thompson, N., et al. The value of comparison. Nature Reviews Microbiology 1, 11 (2003) (link to article)



 Figure 1: Slipped-strand mispairing.
Figure 1: Slipped-strand mispairing.