« Prev Next »
In bacteria, mRNA is translated into protein as soon as it is transcribed. Unlike eukaryotic cells, bacteria do not have a distinct nucleus that separates DNA from ribosomes, so there is no barrier to immediate translation. Indeed, in high-magnification images of bacteria generated by electron microscopy, ribosomes can be seen translating messenger RNAs that are still being transcribed from DNA. This process simply would not work in eukaryotic cells, mainly because eukaryotic RNAs contain introns and exons and must be edited before translation can begin. The eukaryotic nucleus therefore provides a distinct compartment within the cell, allowing transcription and splicing to proceed prior to the beginning of translation. Thus, in eukaryotes, while transcription occurs in the nucleus, translation occurs in the cytoplasm. In other words, eukaryotic transcription and translation are spatially and temporally isolated.
In contrast, bacteria, which have a comparatively streamlined genome, have no need to separate the processes of transcription and translation. In fact, several advantages may be provided by coupling these processes. One advantage involves the energy required for transcription and translation; specifically, the energy needed to drive the process of transcription could be provided by the large-scale expenditure of unstable nucleotide triphosphates during the process of translation. Another advantage to coupled transcription and translation is that it provides a novel mechanism for gene regulation.
Transcriptional Attenuation of the trp Operon
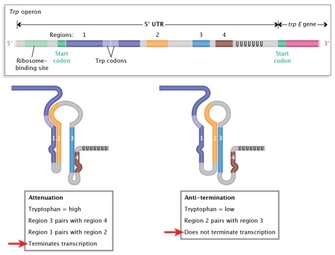
Attenuation, or dampening, of the trp operon is made possible by the fact that the rate of translation influences RNA structure, which in turn influences the rate of transcription. Translation therefore interferes with transcription, making this an example of translation-mediated transcription attenuation. Mechanistically, this kind of attenuation is achieved because special sequences located near the beginning of the transcript, called the leader (trpL), interact to create two possible RNA conformations: one that terminates transcription (the terminator stem), and one that is permissive to transcription (the anti-terminator stem) (Figure 1).
The mechanism by which formation of the terminator stem disrupts continued transcription is now understood. Recent studies have revealed that the formation of this kind of secondary structure in RNA upstream of RNA polymerase can lead to dissolution of the RNA polymerase/DNA complex (Gusarov & Nudler, 1999; Yarnell & Roberts, 1999) and thus a shut-down of further transcription.
Interestingly, the choice between terminator and anti-terminator stem conformations depends on the speed of translation. This adds an additional layer of complexity to the system, as the rate of translation is affected by the availability of trp. Given that trp is an amino acid used to build proteins, the availability of trp will influence the rate at which proteins that contain a lot of trp residues are created. Because the trpL region encodes a trp-rich polypeptide, its translation will be fast when trp is plentiful, and slow when it is not. In turn, quick translation of trpL leads to formation of the terminator stem and attenuation of continued expression of the trp operon. Thus, when trp is plentiful, the coupled processes of transcription and translation respond and shut down.
Transcription, Translation, and Diversity
When it comes to gene regulation, prokaryotes and eukaryotes have evolved the best systems to suit their particular needs. While bacteria and other prokaryotes make use of paired transcription and translation on a variety of levels, eukaryotes have developed a more complex system, with different mechanisms of gene regulation. Indeed, knowledge of the diversity of gene regulatory mechanisms deepens our appreciation of the diversity of nature.
References and Recommended Reading
Gusarov, I., & Nudler, E. The mechanism of intrinsic transcription termination. Molecular Cell 3, 495–504 (1999)
Mandal,
M., & Breaker, R. R. Gene regulation by riboswitches. Nature Reviews
Molecular Cell Biology 5, 451–463 (2004) (link to article)
Merino, E., & Yanofsky, C. Transcription attenuation: A highly conserved regulatory strategy used by bacteria. Trends in Genetics 21, 260–264 (2005)
Yarnell, W. S., & Roberts, J. W. Mechanism of intrinsic transcription termination and anti-termination. Science 284, 611–615 (1999)
Yanofsky, C. Attenuation in the control of expression of bacterial operons. Nature 289, 751–758 (1981) doi:10.1038/289751a0 (link to article)



 Figure 1: Regulation of trp operon transcription.
Figure 1: Regulation of trp operon transcription.


























