« Prev Next »
For most eukaryotic genes (and some prokaryotic ones), the initial RNA that is transcribed from a gene's DNA template must be processed before it becomes a mature messenger RNA (mRNA) that can direct the synthesis of protein. One of the steps in this processing, called RNA splicing, involves the removal or "splicing out" of certain sequences referred to as intervening sequences, or introns. The final mRNA thus consists of the remaining sequences, called exons, which are connected to one another through the splicing process. RNA splicing was initially discovered in the 1970s, overturning years of thought in the field of gene expression.
Early Studies in Bacteria
Gene regulation was first studied most thoroughly in relatively simple bacterial systems. Most bacterial RNA transcripts do not undergo splicing; these transcripts are said to be colinear, with DNA directly encoding them. In other words, there is a one-to-one correspondence of bases between the gene and the mRNA transcribed from the gene (excepting 5′ and 3′ noncoding regions). However, in 1977, several groups of researchers who were working with adenoviruses that infect and replicate in mammalian cells obtained some surprising results. These scientists identified a series of RNA molecules that they termed "mosaics," each of which contained sequences from noncontiguous sites in the viral genome (Berget et al., 1977; Chow et al., 1977). These mosaics were found late in viral infection. Studies of early infection revealed long primary RNA transcripts that contained all of the sequences from the late RNAs, as well as what came to be called the intervening sequences (introns).
Subsequent to the adenoviral discovery, introns were found in many other viral and eukaryotic genes, including those for hemoglobin and immunoglobulin (Darnell, 1978). Splicing of RNA transcripts was then observed in several in vitro systems derived from eukaryotic cells, including removal of introns from transfer RNA in yeast cell-free extracts (Knapp et al., 1978). These observations solidified the hypothesis that splicing of large initial transcripts did, in fact, yield the mature mRNA. Other hypotheses proposed that the DNA template in some way looped or assumed a secondary structure that allowed transcription from noncontiguous regions (Darnell, 1978).
How Splicing Occurs
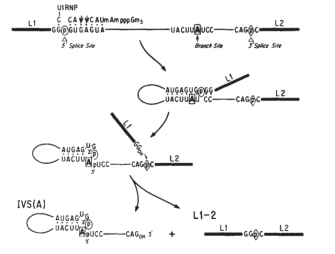
Splicing occurs in several steps and is catalyzed by small nuclear ribonucleoproteins (snRNPs, commonly pronounced "snurps"). First, the pre-mRNA is cleaved at the 5′ end of the intron following the attachment of a snRNP called U1 to its complementary sequence within the intron. The cut end then attaches to the conserved branch point region downstream through pairing of guanine and adenine nucleotides from the 5′ end and the branch point, respectively, to form a looped structure known as a lariat (Figure 1). The bonding of the guanine and adenine bases takes place via a chemical reaction known as transesterification, in which a hydroxyl (OH) group on a carbon atom of the adenine "attacks" the bond of the guanine nucleotide at the splice site. The guanine residue is thus cleaved from the RNA strand and forms a new bond with the adenine.
Next, the snRNPs U2 and U4/U6 appear to contribute to positioning of the 5′ end and the branch point in proximity. With the participation of U5, the 3′ end of the intron is brought into proximity, cut, and joined to the 5′ end. This step occurs by transesterification; in this case, an OH group at the 3′ end of the exon attacks the phosphodiester bond at the 3′ splice site. The adjoining exons are covalently bound, and the resulting lariat is released with U2, U5, and U6 bound to it.
In addition to consensus sequences at their splice sites, eukaryotic genes with long introns also contain exonic splicing enhancers (ESEs). These sequences, which help position the splicing apparatus, are found in the exons of genes and bind proteins that help recruit splicing machinery to the correct site. Most splicing occurs between exons on a single RNA transcript, but occasionally trans-splicing occurs, in which exons on different pre-mRNAs are ligated together.
The splicing process occurs in cellular machines called spliceosomes, in which the snRNPs are found along with additional proteins. The primary variety of spliceosome is one of the most plentiful structures in the cell, and recently, a secondary type of spliceosome has been identified that processes a minor category of introns. These introns are referred to as U12-type introns because they depend upon the action of a snRNP called U12 (the common introns described above are called U2-type introns). The role of U12-type introns is not yet defined, but their persistence throughout evolution and conservation between homologous genes of widely divergent species suggests an important functional basis (Patel & Steitz, 2003).
Self-Splicing and Alternative Splicing
Some RNA molecules have the capacity to splice themselves; the initial discovery of this self-splicing ability in the protozoan Tetrahymena thermophila was recognized with the Nobel Prize in 1989. The self-splicing introns found in T. thermophila are now referred to as Group I introns; this class also includes other protozoan ribosomal RNA genes, some fungal mitochondrial genes, and some phage genes. Group I introns all fold into a complex secondary structure with nine loops and employ transesterification reactions as described above. On the other hand, Group II self-splicing introns are found in mitochondrial genes and are excised by a mechanism that bears similarities to pre-mRNA splicing, including the production of lariats. For this reason, it has been proposed that perhaps pre-mRNA introns and splicing mechanisms evolved from the Group II introns.
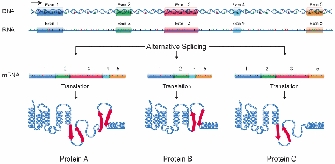
Early in the course of splicing research, yet another surprising discovery was made; specifically, researchers noticed that not only was pre-mRNA punctuated by introns that needed to be excised, but also that alternative patterns of splicing within a single pre-mRNA molecule could yield different functional mRNAs (Figure 2; Berget et al. 1977). The first example of alternative splicing was defined in the adenovirus in 1977 and demonstrated that one pre-mRNA molecule could be spliced at different junctions to result in a variety of mature mRNA molecules, each containing different combinations of exons.
Shortly afterward, alternative splicing was found to occur in cellular genes as well, with the first example identified in the IgM gene, a member of the immunoglobulin superfamily (Early et al., 1980). Another example of a gene with an impressive number of alternative splicing patterns is the Dscam gene from Drosophila, which is involved in guiding embryonic nerves to their targets during formation of the fly's nervous system. Examination of the Dscam sequence reveals such a large number of introns that differential splicing could, in theory, create a staggering 38,000 different mRNAs. This ability to create so many mRNAs may provide the diversity necessary for forming a complex structure such as the nervous system (Schmucker et al., 2000). In fact, the existence of multiple mRNA transcripts within single genes may account for the complexity of some organisms, such as humans, that have relatively few genes (approximately 20,000). For example, work from Wang et al. (2008) suggests that more than 90% of human genes are alternatively spliced.
The Past and Future of Introns
The existence of introns and differential splicing helps explain how new genes are created during evolution. Splicing makes genes more "modular," allowing new combinations of exons to be created during evolution. Furthermore, new exons can be inserted into old introns, creating new proteins without disrupting the function of the old gene.
Our knowledge of RNA splicing is quite new. Nonetheless, because nearly all eukaryotes have introns and share mechanisms of RNA splicing, splicing itself must be quite ancient. Proponents of the "intron-early" theory suggest that all organisms (including prokaryotes) at one time had introns in their genome but subsequently lost these elements, while "intron-late" supporters believe that the restriction of introns to eukaryotes suggests a more recent introduction (Roy & Gilbert, 2006). There is no apparent pattern in which eukaryotes have introns, and that makes it difficult for researchers to make predictions about how introns were gained or lost through evolution. What is clear, however, is that introns and splicing have clearly played a significant role in evolution, and scientists are only beginning to discover the nature of that role.
References and Recommended Reading
Berget, S. M., et al. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proceedings of the National Academy of Sciences 74, 3171–3175 (1977)
Chow, L. T., et al. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977)
Darnell, J. E., Jr. Implications of RNA–RNA splicing in evolution of eukaryotic cells. Science 202, 1257–1260 (1978) doi:10.1126/science.364651
Early, P., et al. Two mRNAs can be produced from a single immunoglobulin chain by alternative RNA processing pathways. Cell 20, 313–319 (1980)
Knapp, G., et al. Transcription and processing of intervening sequences in yeast tRNA genes. Cell 14, 221–236 (1978)
Konarska, M. M., et al. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313, 552–557 (1984) doi: 10.1038/313552a0 (link to article)
Patel, A. A., & Steitz, J. A. Splicing double: Insights from the second spliceosome. Nature 4, 960–970 (2003) doi:10.1038/nrm1259 (link to article)
Pierce, B. A. Genetics: A Conceptual Approach, 2nd ed. (New York, Freeman, 2000)
Roy, S. W., & Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles, and progress. Nature Reviews Genetics 7, 211–221 (2006) doi: 10.1038/nrg1807 (link to article)
Schmucker, D., et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 (2000)



 Figure 1: Pre-mRNA splicing.
Figure 1: Pre-mRNA splicing.